Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101634 |
| Name | oriT_CP4|p3 |
| Organism | Enterococcus faecium strain CP4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAMWMK010000146 (26367..26466 [+], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 13..18, 32..37 (AAAGGG..CCCTTT) |
| Location of nic site | 94..95 |
| Conserved sequence flanking the nic site |
GCTTGCAGTA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
>oriT_CP4|p3
CCGAAGGGGATGAAAGGGAACGTGTAACTTGCCCTTTTCCAAGCAACTTTGTCATACAAGACACTTCGTTATTGTATGACAAAGTGAAAGCTTGCAGTAT
CCGAAGGGGATGAAAGGGAACGTGTAACTTGCCCTTTTCCAAGCAACTTTGTCATACAAGACACTTCGTTATTGTATGACAAAGTGAAAGCTTGCAGTAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file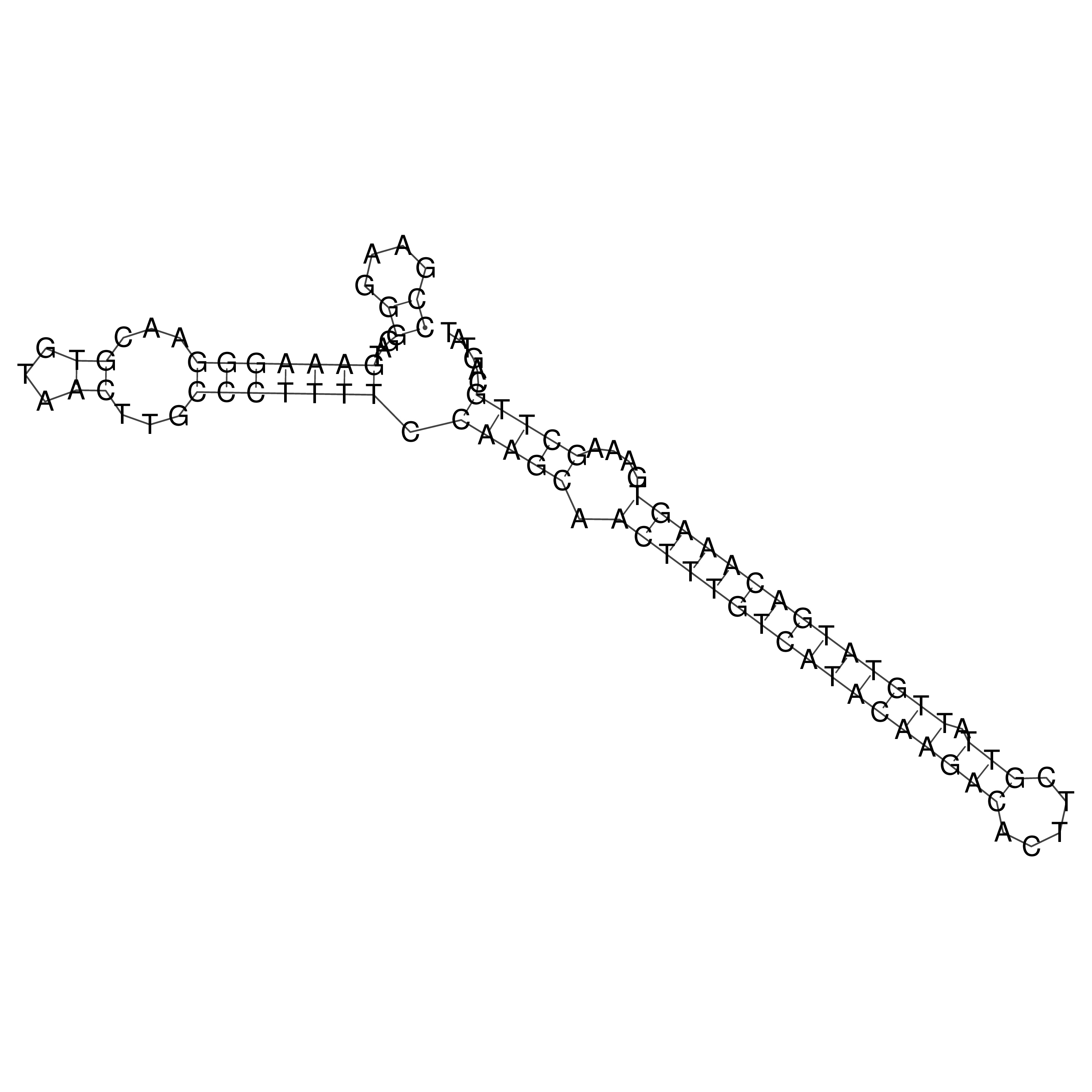
Relaxase
| ID | 1430 | GenBank | WP_258863622 |
| Name | Relaxase_M3X98_RS15375_CP4|p3 |
UniProt ID | _ |
| Length | 342 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 342 a.a. Molecular weight: 40259.66 Da Isoelectric Point: 10.2510
>WP_258863622.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterococcus]
MHNKTQERNQVLRITQSFALDELSPKNTQDWQRANDLGVALAEKLYPQHQTAVYTHLDGQNHVLHNHILV
NKVNLETGKKLREVPGKTVEKARVINDRLAQQQHWRILPPPQERFSETEQDLVAKQKYSYMDDLRTRINE
AMVDISVRSYKDLRNILEQKSIIFQERVQSLSYAFLDVNNKKRRAKGARLGTDFEKETIFNELENRARQQ
QPRPIEPVKQRELATTRLMQDTQQTEPSLERRESTTRGIEQQLEQRKPGIARLIDTIQHLRDQLPGITQL
VKKHLTYAKEALFDAFEKRFSFDMAQHKQVQRKKIQQDLEQRTQEQAMPKFQTPTQDRGLSL
MHNKTQERNQVLRITQSFALDELSPKNTQDWQRANDLGVALAEKLYPQHQTAVYTHLDGQNHVLHNHILV
NKVNLETGKKLREVPGKTVEKARVINDRLAQQQHWRILPPPQERFSETEQDLVAKQKYSYMDDLRTRINE
AMVDISVRSYKDLRNILEQKSIIFQERVQSLSYAFLDVNNKKRRAKGARLGTDFEKETIFNELENRARQQ
QPRPIEPVKQRELATTRLMQDTQQTEPSLERRESTTRGIEQQLEQRKPGIARLIDTIQHLRDQLPGITQL
VKKHLTYAKEALFDAFEKRFSFDMAQHKQVQRKKIQQDLEQRTQEQAMPKFQTPTQDRGLSL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 2078 | GenBank | NZ_JAMWMK010000146 |
| Plasmid name | CP4|p3 | Incompatibility group | - |
| Plasmid size | 35098 bp | Coordinate of oriT [Strand] | 26367..26466 [+] |
| Host baterium | Enterococcus faecium strain CP4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA1 |