Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101600 |
| Name | oriT_2020CK-00188|unnamed1 |
| Organism | Escherichia coli strain 2020CK-00188 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_ABACVG020000002 (26149..26251 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 64..70, 73..79 (CCACCGT..ACGGTGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_2020CK-00188|unnamed1
AGCCTCGCAGAGCAGGATTCCCGTTGAGCAGCCCAGCTGCGAATAAGGGAAGGTAAAGTTGGACCACCGTTTACGGTGGGCCTACTTTACATATCCTGCCCGG
AGCCTCGCAGAGCAGGATTCCCGTTGAGCAGCCCAGCTGCGAATAAGGGAAGGTAAAGTTGGACCACCGTTTACGGTGGGCCTACTTTACATATCCTGCCCGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file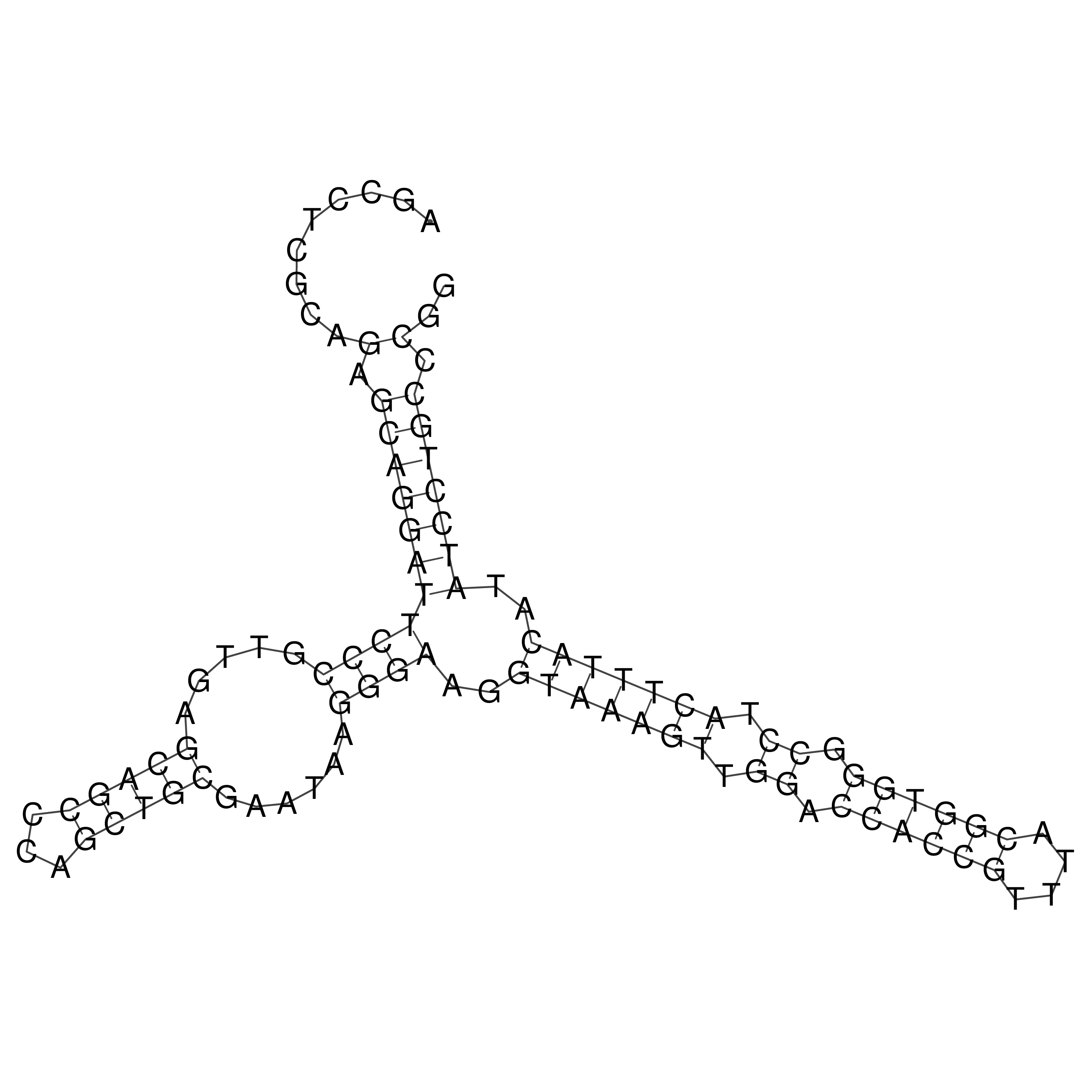
Relaxase
| ID | 1417 | GenBank | WP_050580356 |
| Name | traI_IUB69_RS23280_2020CK-00188|unnamed1 |
UniProt ID | _ |
| Length | 869 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 869 a.a. Molecular weight: 96507.42 Da Isoelectric Point: 10.4435
>WP_050580356.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacteriaceae]
MKNIKKSSFSGLADYITNPQSKQERVGVIKATNCQSQDVQDAVTEVNITQRLNTRSESDKTYHLVISFRP
GEKPSEDVLNAIEESICQSLGFGDHQRLSAVHHDTDNLHIHVAINKINPDTLTIHNPYNDYRVLAESCAY
LEKKYGLEVDNHMPTRSASENRAKDMERHSGIQSLLSWVKAECAEQLLNANSWQELHQAMRQNGLELRQK
GNGFVIAGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRPSPEAIKQRFGSAANVKRPGDRPP
PMRENRQVHLGDVPVMQINSGKRYESKPLGSQGANTTELYGRYQSEQQRLTAARNEALRLAKVKRDRLIE
NAKRSGRLKRATIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKAA
GGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVSR
DAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFADPELEKLRQILTKKAAREREYI
HIPFEKRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTDAGRERSGNG
NATGANEPGVTGRSTSADAGNGYGTSSSRRRTGTGDAVTTAGSQPGRSDDTRTGRSRAAEPNGERGHSKP
NVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPAE
VKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDYT
ARRLSNLKVGESVTLTPSGSVRTTKGRSR
MKNIKKSSFSGLADYITNPQSKQERVGVIKATNCQSQDVQDAVTEVNITQRLNTRSESDKTYHLVISFRP
GEKPSEDVLNAIEESICQSLGFGDHQRLSAVHHDTDNLHIHVAINKINPDTLTIHNPYNDYRVLAESCAY
LEKKYGLEVDNHMPTRSASENRAKDMERHSGIQSLLSWVKAECAEQLLNANSWQELHQAMRQNGLELRQK
GNGFVIAGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRPSPEAIKQRFGSAANVKRPGDRPP
PMRENRQVHLGDVPVMQINSGKRYESKPLGSQGANTTELYGRYQSEQQRLTAARNEALRLAKVKRDRLIE
NAKRSGRLKRATIKLLKGPGVNKRYLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWADWLQFKAA
GGDDEALKTLRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDGGKRLDVSR
DAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFADPELEKLRQILTKKAAREREYI
HIPFEKRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTDAGRERSGNG
NATGANEPGVTGRSTSADAGNGYGTSSSRRRTGTGDAVTTAGSQPGRSDDTRTGRSRAAEPNGERGHSKP
NVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLQGNVSDQLEHKTAERADQLRRDMAGAGGITTPAE
VKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIVVMPVSDYT
ARRLSNLKVGESVTLTPSGSVRTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1074 | GenBank | WP_024252940 |
| Name | t4cp2_IUB69_RS23270_2020CK-00188|unnamed1 |
UniProt ID | _ |
| Length | 648 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 648 a.a. Molecular weight: 71684.76 Da Isoelectric Point: 8.1395
>WP_024252940.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPTKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAVAANDDASNEAIAI
MRKPNNAVGPQVRNKKKTARGKLIPAMAITSLAAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHDRLPDAFFAAGSVGATVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIKAAGLLGNDEGV
YVGAWEDKNGQLQYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYA
KNLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVL
YKHKNDGTPATLPYVDSIMADPERGAGELWMEMTQYGHVNGENHPVVGAAGRDMMDRPEEEAGSVLSTAK
SYLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERV
NNNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLAMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQD
INQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTM
QEVQRPLLTVDECLRMPGPTKNAEGLITERGDMVVYVAGFPAIYGKQPLFFQDEIFSLRASVPEPKTTDR
IRAAVAANDDASNEAIAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 44944..50807
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IUB69_RS23395 (IUB69_023335) | 40105..40302 | - | 198 | WP_024252921 | DUF905 family protein | - |
| IUB69_RS23400 (IUB69_023340) | 40392..40976 | - | 585 | WP_024252920 | hypothetical protein | - |
| IUB69_RS23405 (IUB69_023345) | 40973..41221 | - | 249 | WP_024252919 | antirestriction protein ArdR | - |
| IUB69_RS23410 (IUB69_023350) | 41221..41529 | - | 309 | WP_003019666 | HigA family addiction module antitoxin | - |
| IUB69_RS23415 (IUB69_023355) | 41538..41837 | - | 300 | WP_024252918 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| IUB69_RS23420 (IUB69_023360) | 42850..43728 | - | 879 | WP_032194784 | plasmid replication initiator TrfA | - |
| IUB69_RS23425 (IUB69_023365) | 43809..44180 | - | 372 | WP_024252967 | single-stranded DNA-binding protein | - |
| IUB69_RS23430 (IUB69_023370) | 44286..44690 | + | 405 | WP_024252966 | transcriptional regulator | - |
| IUB69_RS23435 (IUB69_023375) | 44944..45831 | + | 888 | WP_024252965 | P-type conjugative transfer ATPase TrbB | virB11 |
| IUB69_RS23440 (IUB69_023380) | 45945..46328 | + | 384 | WP_024252964 | TrbC/VirB2 family protein | virB2 |
| IUB69_RS23445 (IUB69_023385) | 46331..46657 | + | 327 | WP_270098195 | conjugal transfer protein TrbD | virB3 |
| IUB69_RS23450 (IUB69_023390) | 46654..49197 | + | 2544 | WP_270098197 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| IUB69_RS23455 (IUB69_023395) | 49194..49901 | + | 708 | WP_024252961 | VirB8/TrbF family protein | virB8 |
| IUB69_RS23460 (IUB69_023400) | 49917..50807 | + | 891 | WP_024252960 | P-type conjugative transfer protein TrbG | virB9 |
| IUB69_RS23465 (IUB69_023405) | 50810..51262 | + | 453 | WP_032194780 | conjugal transfer protein TrbH | - |
Host bacterium
| ID | 2044 | GenBank | NZ_ABACVG020000002 |
| Plasmid name | 2020CK-00188|unnamed1 | Incompatibility group | IncP1 |
| Plasmid size | 52406 bp | Coordinate of oriT [Strand] | 26149..26251 [+] |
| Host baterium | Escherichia coli strain 2020CK-00188 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |