Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101571 |
| Name | oriT_pMFDS1013836 |
| Organism | Escherichia coli strain MFDS1013836 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAQMUU010000004 (5042..5165 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 41..48, 61..68 (AAAAACAA..TTGTTTTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGTTTTTTTAATCATTAGTTTATGTTCTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file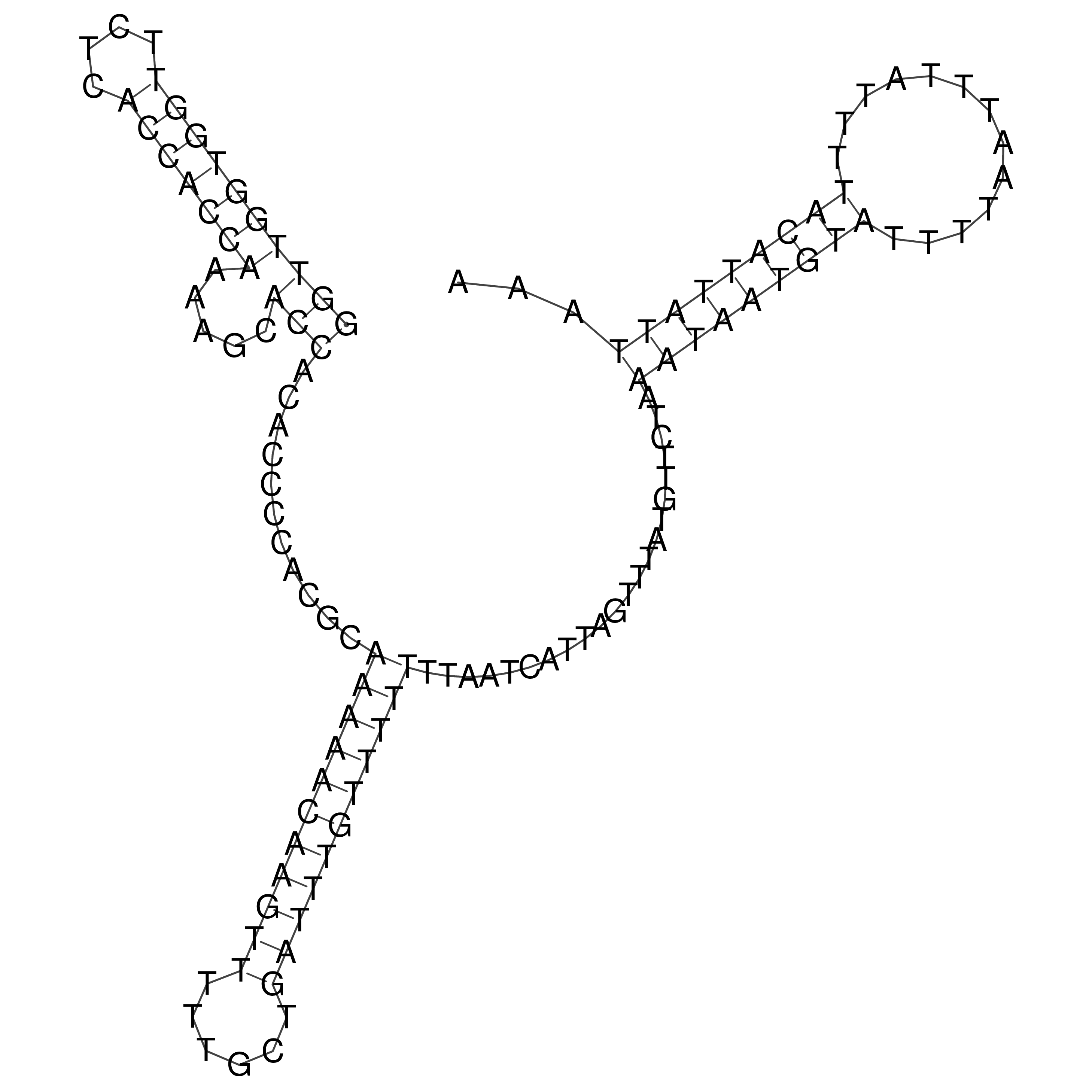
Relaxase
| ID | 1393 | GenBank | WP_064578954 |
| Name | traI_V6M02_RS27885_pMFDS1013836 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192244.09 Da Isoelectric Point: 5.9188
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGRNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIEAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERVRAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMIAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGRPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSNSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFMALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYETEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPNFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVELPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAGQERERAVTSEREAALPESVLREPQREREAVREVVRENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 521 | GenBank | WP_040073112 |
| Name | WP_040073112_pMFDS1013836 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14556.62 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSIEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 522 | GenBank | WP_001254386 |
| Name | WP_001254386_pMFDS1013836 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 1033 | GenBank | WP_001064245 |
| Name | traC_V6M02_RS27780_pMFDS1013836 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99159.81 Da Isoelectric Point: 5.8486
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNVADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1034 | GenBank | WP_062873762 |
| Name | traD_V6M02_RS27870_pMFDS1013836 |
UniProt ID | _ |
| Length | 717 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 717 a.a. Molecular weight: 81434.90 Da Isoelectric Point: 5.0485
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAASLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPDMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPVSPAI
NDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPDIQQQMQRREEVN
INVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4470..30607
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| V6M02_RS27655 | 1..382 | + | 382 | Protein_0 | hypothetical protein | - |
| V6M02_RS27660 | 437..871 | + | 435 | WP_000845953 | conjugation system SOS inhibitor PsiB | - |
| V6M02_RS27665 | 868..1630 | + | 763 | Protein_2 | plasmid SOS inhibition protein A | - |
| V6M02_RS27670 | 1599..1787 | - | 189 | WP_032084246 | hypothetical protein | - |
| V6M02_RS27675 | 1809..1958 | + | 150 | Protein_4 | plasmid maintenance protein Mok | - |
| V6M02_RS27680 | 1900..2025 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| V6M02_RS27685 | 2245..2475 | + | 231 | WP_074014914 | hypothetical protein | - |
| V6M02_RS27690 | 2473..2646 | - | 174 | Protein_7 | hypothetical protein | - |
| V6M02_RS27695 | 2716..2922 | + | 207 | WP_000547968 | hypothetical protein | - |
| V6M02_RS27700 | 2947..3233 | + | 287 | Protein_9 | hypothetical protein | - |
| V6M02_RS27705 | 3352..4173 | + | 822 | WP_074014913 | DUF932 domain-containing protein | - |
| V6M02_RS27710 | 4470..5117 | - | 648 | WP_123007297 | transglycosylase SLT domain-containing protein | virB1 |
| V6M02_RS27715 | 5394..5777 | + | 384 | WP_040073112 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| V6M02_RS27720 | 5967..6653 | + | 687 | WP_000332492 | PAS domain-containing protein | - |
| V6M02_RS27725 | 6747..6974 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| V6M02_RS27730 | 7008..7373 | + | 366 | WP_000340270 | type IV conjugative transfer system pilin TraA | - |
| V6M02_RS27735 | 7388..7699 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| V6M02_RS27740 | 7721..8287 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| V6M02_RS27745 | 8274..9001 | + | 728 | Protein_18 | type-F conjugative transfer system secretin TraK | - |
| V6M02_RS27750 | 9001..10428 | + | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| V6M02_RS27755 | 10418..11008 | + | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| V6M02_RS27760 | 10995..11192 | + | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| V6M02_RS27765 | 11204..11454 | + | 251 | Protein_22 | conjugal transfer protein TrbG | - |
| V6M02_RS27770 | 11451..11966 | + | 516 | WP_332829757 | type IV conjugative transfer system lipoprotein TraV | traV |
| V6M02_RS27775 | 12101..12322 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| V6M02_RS27780 | 12482..15109 | + | 2628 | WP_001064245 | type IV secretion system protein TraC | virb4 |
| V6M02_RS27785 | 15106..15492 | + | 387 | WP_032082946 | type-F conjugative transfer system protein TrbI | - |
| V6M02_RS27790 | 15489..16121 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| V6M02_RS27795 | 16118..17110 | + | 993 | WP_000830183 | conjugal transfer pilus assembly protein TraU | traU |
| V6M02_RS27800 | 17119..17757 | + | 639 | WP_000777695 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| V6M02_RS27805 | 17754..19562 | + | 1809 | WP_032082947 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| V6M02_RS27810 | 19589..19846 | + | 258 | WP_000864318 | conjugal transfer protein TrbE | - |
| V6M02_RS27815 | 19839..20582 | + | 744 | WP_032082948 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| V6M02_RS27820 | 20598..20945 | + | 348 | WP_115429732 | conjugal transfer protein TrbA | - |
| V6M02_RS27825 | 20947..21222 | - | 276 | WP_001513501 | hypothetical protein | - |
| V6M02_RS27830 | 21303..21587 | + | 285 | WP_000624108 | type-F conjugative transfer system pilin chaperone TraQ | - |
| V6M02_RS27835 | 21574..22119 | + | 546 | WP_032082911 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| V6M02_RS27840 | 22049..22390 | + | 342 | WP_001442101 | P-type conjugative transfer protein TrbJ | - |
| V6M02_RS27845 | 22377..22769 | + | 393 | WP_032082924 | F-type conjugal transfer protein TrbF | - |
| V6M02_RS27850 | 22756..24129 | + | 1374 | WP_000944319 | conjugal transfer pilus assembly protein TraH | traH |
| V6M02_RS27855 | 24126..26951 | + | 2826 | WP_032082912 | conjugal transfer mating-pair stabilization protein TraG | traG |
| V6M02_RS27860 | 26948..27457 | + | 510 | WP_032082913 | conjugal transfer entry exclusion protein TraS | - |
| V6M02_RS27865 | 27471..28202 | + | 732 | WP_024187508 | conjugal transfer complement resistance protein TraT | - |
| V6M02_RS27870 | 28454..30607 | + | 2154 | WP_062873762 | type IV conjugative transfer system coupling protein TraD | virb4 |
| V6M02_RS27875 | 30616..31014 | - | 399 | WP_000911317 | type II toxin-antitoxin system VapC family toxin | - |
| V6M02_RS27880 | 31014..31241 | - | 228 | WP_000450532 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 2015 | GenBank | NZ_JAQMUU010000004 |
| Plasmid name | pMFDS1013836 | Incompatibility group | IncFII |
| Plasmid size | 101631 bp | Coordinate of oriT [Strand] | 5042..5165 [+] |
| Host baterium | Escherichia coli strain MFDS1013836 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | estIa |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |