Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101556 |
| Name | oriT_FDAARGOS_950|unnamed |
| Organism | Hafnia alvei strain FDAARGOS_950 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAEDZV010000012 (21201..21286 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
>oriT_FDAARGOS_950|unnamed
TAAGGGACAAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGCCCCTACAAAACGTCTTGGTCAGGGCGAAGCCCCGACACCCC
TAAGGGACAAGATGTGTTTTGTAGCACCGCCTGCACGCAGTCGGCCCCTACAAAACGTCTTGGTCAGGGCGAAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file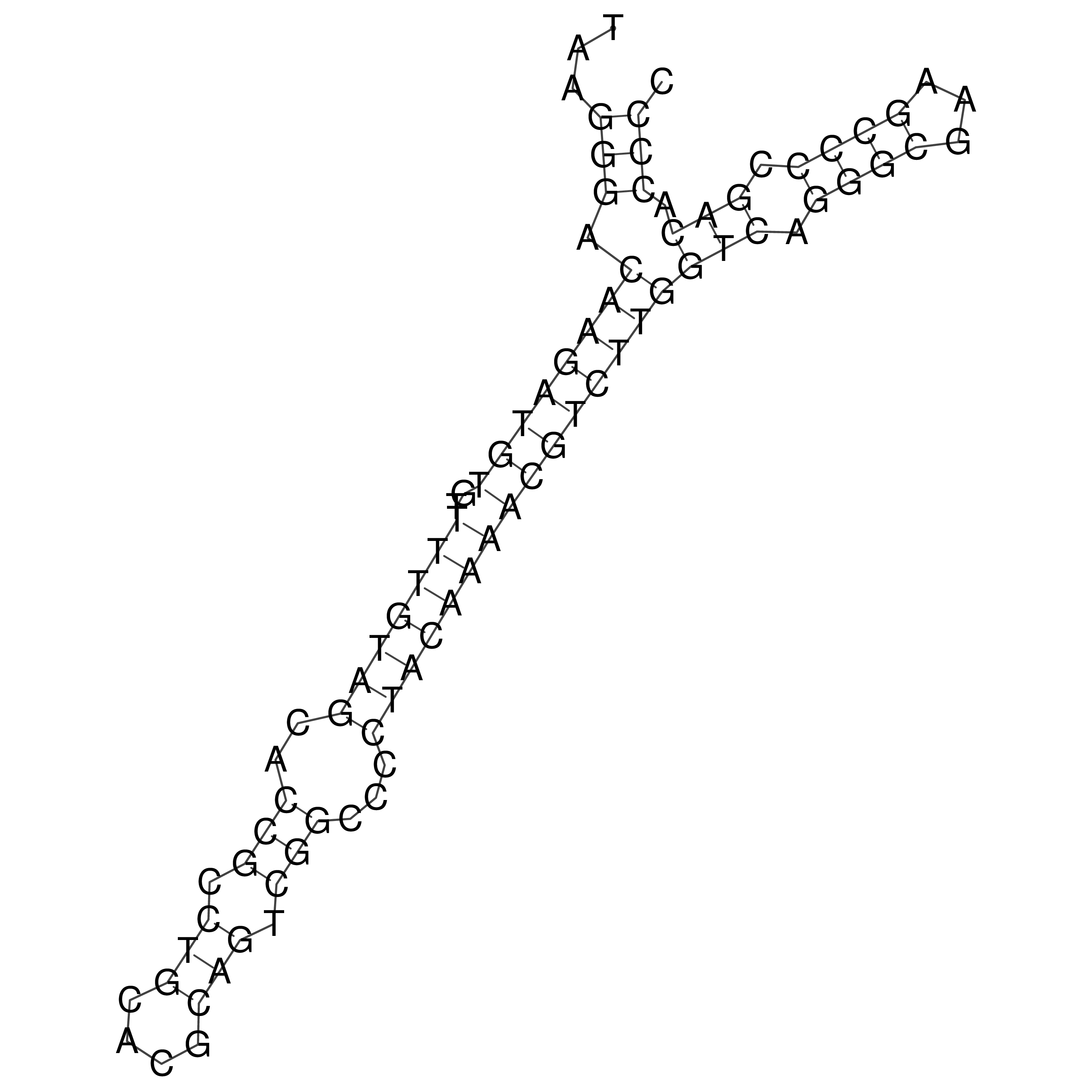
Relaxase
| ID | 1379 | GenBank | WP_103008360 |
| Name | traI_I6H07_RS22585_FDAARGOS_950|unnamed |
UniProt ID | _ |
| Length | 664 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 664 a.a. Molecular weight: 76808.31 Da Isoelectric Point: 6.6708
>WP_103008360.1 TraI/MobA(P) family conjugative relaxase [Hafnia alvei]
MIGRIPPKRRDGKSSFLKLVSYVVIRDEDKPDMPLEPEHPDWRRPKSKDDLFNRLVDYVSRSGDEGAMHT
LNIDDDGRQRVLFDGVLCETNNFSLATAAVEMNAVASQNTRCKDPVFHYILSWPTSDNPNQDEVFDSVRH
TLKSFGMDEHQYVAAIHTDTNNIHCHIAVNRVHPVTYDTADDSFTRLKLQRACRELELKYDWTPTNGCYV
VDENKQIVYKKRDEPPVPQGSVAMEYYADQESLHSYAMRTCSDDLEKLIVKDELTWFELHKLLVKQGLRL
DKKGKGLAIYSLDEKDITPIKASHLHPDLTLNCLEDDLGPFVRMEDVGRYTLDEGDTSEDALIHMYRYEP
HLHKRDEGARASRREARAVARRELFDRYKKYSNGYKRPKMEGTEATTQFKALSNRYAWKKEQVRFVFDDP
LIRKAVYRVLEAERQKEFESLKAQIKQERNAFYSHPANRKLTKQEWVGQQALKQDQAAISCLRGWSYRYK
RNALTPTLSDNAIVCAVADDIKPYNVQSYETSVNRDGTIQYKQDGIVQIQDKGNSIEIADPYIQNGQHIA
GAMVIAEEKSGEKLLFSGETEFINRACSLVPWFNSDGEKPLTLTDPQQRVMAGYDKPEVRDDKFVATHRI
VDEETYLASLERNEVNDNRERHDLKSKKNSYRPR
MIGRIPPKRRDGKSSFLKLVSYVVIRDEDKPDMPLEPEHPDWRRPKSKDDLFNRLVDYVSRSGDEGAMHT
LNIDDDGRQRVLFDGVLCETNNFSLATAAVEMNAVASQNTRCKDPVFHYILSWPTSDNPNQDEVFDSVRH
TLKSFGMDEHQYVAAIHTDTNNIHCHIAVNRVHPVTYDTADDSFTRLKLQRACRELELKYDWTPTNGCYV
VDENKQIVYKKRDEPPVPQGSVAMEYYADQESLHSYAMRTCSDDLEKLIVKDELTWFELHKLLVKQGLRL
DKKGKGLAIYSLDEKDITPIKASHLHPDLTLNCLEDDLGPFVRMEDVGRYTLDEGDTSEDALIHMYRYEP
HLHKRDEGARASRREARAVARRELFDRYKKYSNGYKRPKMEGTEATTQFKALSNRYAWKKEQVRFVFDDP
LIRKAVYRVLEAERQKEFESLKAQIKQERNAFYSHPANRKLTKQEWVGQQALKQDQAAISCLRGWSYRYK
RNALTPTLSDNAIVCAVADDIKPYNVQSYETSVNRDGTIQYKQDGIVQIQDKGNSIEIADPYIQNGQHIA
GAMVIAEEKSGEKLLFSGETEFINRACSLVPWFNSDGEKPLTLTDPQQRVMAGYDKPEVRDDKFVATHRI
VDEETYLASLERNEVNDNRERHDLKSKKNSYRPR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1012 | GenBank | WP_103008353 |
| Name | trbC_I6H07_RS22545_FDAARGOS_950|unnamed |
UniProt ID | _ |
| Length | 731 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 731 a.a. Molecular weight: 83005.71 Da Isoelectric Point: 7.7658
>WP_103008353.1 F-type conjugative transfer protein TrbC [Hafnia alvei]
MPNDNTQPVDKVRVRQTTQSTWIDDYLLAPVTVQAGLVVILILTFLRTWVLIPGVFATAIWLLTFFGQLF
RMPLRMPKDIGGLDMSTERENAYEFKGPMGLFRFTRRRLSWDKSAGIMCLGHARRRFLGRELWLTLEDCL
RHLQLLATTGSGKTEALLSIQLNSICMGRGMTFSDGKAETRLAYAIWSLMRRYGQEDNYYVLNFLNGGRD
RFDELLGNDRTRPQSNSINFFGDATATFIIQLMESLLPQVGASEAGWQDKAKPMLYGLVYALYYKCKKES
IRLSQSMIQKHLPLKKMVELYIEGKQNGWHDEALNALESYLNNLAGFRMELVSKPSEWDQGVYDQHGFLI
QQFSRMLAMFNDVYGHIFATDAGDIDIRDVLHNDRTLVVLIPALELSKSESANLGKLYISAKRMVISRDL
GYQLEGKRKDVLIAKKFANPFPYPDIHDELGSYFAPGMDDLAAQMRSLGMMLVISAQDIQRFIAQFKGEY
QTVNANMLVKWFMALQDEKDTFELAKATAGKDYYAELGEMKRTPGTVSSSYEEANTTYIREKNRISLDEL
KELNPGEGFISFKSALVPSSAIYIPDDEKLSSKLELRINRFIDIGSPTEADLIAQNPHLSRLLPPSEMEV
SMVLQRTDEIVAGSETHTIPPLMDPILARLMKVAMDLDNRCDISYSPTQRGILLFEAARDVLNKRGKKWF
TVKTQPNPLKVSDAMARRLAKQSTAFMSQGH
MPNDNTQPVDKVRVRQTTQSTWIDDYLLAPVTVQAGLVVILILTFLRTWVLIPGVFATAIWLLTFFGQLF
RMPLRMPKDIGGLDMSTERENAYEFKGPMGLFRFTRRRLSWDKSAGIMCLGHARRRFLGRELWLTLEDCL
RHLQLLATTGSGKTEALLSIQLNSICMGRGMTFSDGKAETRLAYAIWSLMRRYGQEDNYYVLNFLNGGRD
RFDELLGNDRTRPQSNSINFFGDATATFIIQLMESLLPQVGASEAGWQDKAKPMLYGLVYALYYKCKKES
IRLSQSMIQKHLPLKKMVELYIEGKQNGWHDEALNALESYLNNLAGFRMELVSKPSEWDQGVYDQHGFLI
QQFSRMLAMFNDVYGHIFATDAGDIDIRDVLHNDRTLVVLIPALELSKSESANLGKLYISAKRMVISRDL
GYQLEGKRKDVLIAKKFANPFPYPDIHDELGSYFAPGMDDLAAQMRSLGMMLVISAQDIQRFIAQFKGEY
QTVNANMLVKWFMALQDEKDTFELAKATAGKDYYAELGEMKRTPGTVSSSYEEANTTYIREKNRISLDEL
KELNPGEGFISFKSALVPSSAIYIPDDEKLSSKLELRINRFIDIGSPTEADLIAQNPHLSRLLPPSEMEV
SMVLQRTDEIVAGSETHTIPPLMDPILARLMKVAMDLDNRCDISYSPTQRGILLFEAARDVLNKRGKKWF
TVKTQPNPLKVSDAMARRLAKQSTAFMSQGH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 923..12360
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I6H07_RS22495 (I6H07_22515) | 491..886 | + | 396 | WP_103008346 | hypothetical protein | - |
| I6H07_RS22500 (I6H07_22520) | 923..1531 | + | 609 | WP_103008347 | hypothetical protein | traT |
| I6H07_RS22505 (I6H07_22525) | 1528..4590 | + | 3063 | WP_103008348 | conjugal transfer protein | traU |
| I6H07_RS22510 (I6H07_22530) | 4608..5828 | + | 1221 | WP_103008349 | conjugal transfer protein TraW | traW |
| I6H07_RS22515 (I6H07_22535) | 5825..6343 | + | 519 | WP_046449685 | conjugal transfer protein TraX | - |
| I6H07_RS22520 (I6H07_22540) | 6394..8526 | + | 2133 | WP_103008350 | DotA/TraY family protein | traY |
| I6H07_RS22525 (I6H07_22545) | 8537..9097 | + | 561 | WP_127908307 | hypothetical protein | - |
| I6H07_RS23590 | 9690..9785 | + | 96 | WP_021542841 | DinQ-like type I toxin DqlB | - |
| I6H07_RS22535 (I6H07_22555) | 9929..11224 | + | 1296 | WP_232353167 | conjugal transfer protein TrbA | trbA |
| I6H07_RS22540 (I6H07_22560) | 11371..12360 | + | 990 | WP_103008352 | thioredoxin fold domain-containing protein | trbB |
| I6H07_RS22545 (I6H07_22565) | 12347..14542 | + | 2196 | WP_103008353 | F-type conjugative transfer protein TrbC | - |
| I6H07_RS22550 (I6H07_22570) | 14682..15185 | + | 504 | WP_103008373 | phospholipase D family protein | - |
| I6H07_RS22555 (I6H07_22575) | 15250..15747 | + | 498 | WP_103008354 | hypothetical protein | - |
| I6H07_RS22560 (I6H07_22580) | 15892..16467 | + | 576 | WP_103008355 | DUF2913 family protein | - |
| I6H07_RS22565 (I6H07_22585) | 16583..16774 | + | 192 | WP_103008356 | hypothetical protein | - |
| I6H07_RS22570 (I6H07_22590) | 16786..17052 | + | 267 | WP_103008357 | PerC family transcriptional regulator | - |
Region 2: 139340..144490
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I6H07_RS23170 (I6H07_23190) | 135135..135671 | + | 537 | WP_103008333 | transcription termination factor NusG | - |
| I6H07_RS23175 (I6H07_23195) | 135880..136536 | + | 657 | WP_232353166 | hypothetical protein | - |
| I6H07_RS23180 (I6H07_23200) | 136599..136802 | + | 204 | WP_103008334 | TraR/DksA C4-type zinc finger protein | - |
| I6H07_RS23185 (I6H07_23205) | 136816..137010 | + | 195 | WP_046449659 | hypothetical protein | - |
| I6H07_RS23190 (I6H07_23210) | 137537..137950 | + | 414 | WP_127908306 | hypothetical protein | - |
| I6H07_RS23195 (I6H07_23215) | 138001..138987 | + | 987 | WP_186277457 | TcpQ domain-containing protein | - |
| I6H07_RS23200 (I6H07_23220) | 138987..139343 | + | 357 | WP_103008338 | type IV pilus biogenesis protein PilM | - |
| I6H07_RS23205 (I6H07_23225) | 139340..139804 | + | 465 | WP_064575603 | hypothetical protein | traL |
| I6H07_RS23210 (I6H07_23230) | 139868..140545 | + | 678 | WP_103008339 | DotI/IcmL/TraM family protein | traM |
| I6H07_RS23215 (I6H07_23235) | 140542..141579 | + | 1038 | WP_103008340 | DotH/IcmK family type IV secretion protein | traN |
| I6H07_RS23220 (I6H07_23240) | 141579..142781 | + | 1203 | WP_103008341 | conjugal transfer protein TraO | traO |
| I6H07_RS23225 (I6H07_23245) | 142778..143536 | + | 759 | WP_103008342 | hypothetical protein | traP |
| I6H07_RS23230 (I6H07_23250) | 143546..144076 | + | 531 | WP_103008343 | conjugal transfer protein TraQ | traQ |
| I6H07_RS23235 (I6H07_23255) | 144092..144490 | + | 399 | WP_046449680 | DUF6750 family protein | traR |
| I6H07_RS23240 (I6H07_23260) | 144523..145098 | - | 576 | WP_103008344 | hypothetical protein | - |
Host bacterium
| ID | 2000 | GenBank | NZ_JAEDZV010000012 |
| Plasmid name | FDAARGOS_950|unnamed | Incompatibility group | - |
| Plasmid size | 145098 bp | Coordinate of oriT [Strand] | 21201..21286 [-] |
| Host baterium | Hafnia alvei strain FDAARGOS_950 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |