Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101553 |
| Name | oriT_L2M4-08|unnamed1 |
| Organism | Escherichia coli strain L2M4-08 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAPVEN010000034 (49635..49735 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_L2M4-08|unnamed1
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file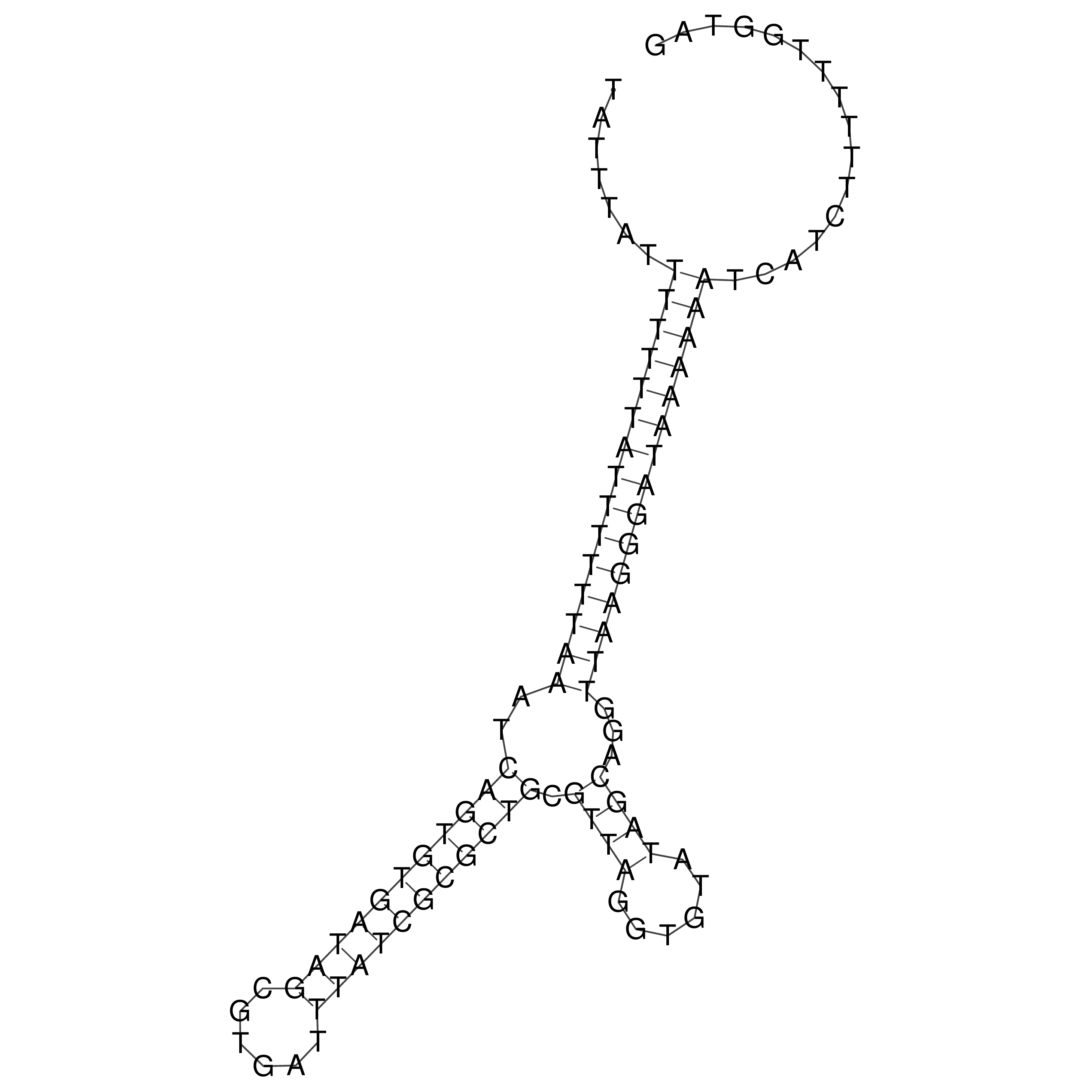
Relaxase
| ID | 1376 | GenBank | WP_000884352 |
| Name | mobF_O3I04_RS19360_L2M4-08|unnamed1 |
UniProt ID | A8R732 |
| Length | 1078 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120163.41 Da Isoelectric Point: 6.5766
>WP_000884352.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A8R732 |
Auxiliary protein
| ID | 496 | GenBank | WP_000706094 |
| Name | WP_000706094_L2M4-08|unnamed1 |
UniProt ID | B6UZ32 |
| Length | 96 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 96 a.a. Molecular weight: 10829.54 Da Isoelectric Point: 8.5040
>WP_000706094.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B6UZ32 |
T4CP
| ID | 1009 | GenBank | WP_000342687 |
| Name | t4cp2_O3I04_RS19365_L2M4-08|unnamed1 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57774.92 Da Isoelectric Point: 9.6551
>WP_000342687.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacteriaceae]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARILADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARILADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14616..24941
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| O3I04_RS19160 (O3I04_19085) | 10467..11900 | - | 1434 | WP_001288435 | DNA cytosine methyltransferase | - |
| O3I04_RS19165 (O3I04_19090) | 11994..12266 | + | 273 | Protein_13 | IS1 family transposase | - |
| O3I04_RS19170 (O3I04_19095) | 12286..12483 | - | 198 | WP_000844102 | hypothetical protein | - |
| O3I04_RS19175 (O3I04_19100) | 12488..12997 | - | 510 | WP_000893479 | restriction endonuclease | - |
| O3I04_RS19180 (O3I04_19105) | 13185..13496 | - | 312 | WP_001452736 | hypothetical protein | - |
| O3I04_RS19185 (O3I04_19110) | 13532..13846 | - | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| O3I04_RS19190 (O3I04_19115) | 13809..14186 | - | 378 | WP_000504251 | hypothetical protein | - |
| O3I04_RS19195 (O3I04_19120) | 14202..14573 | - | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| O3I04_RS19200 (O3I04_19125) | 14616..15350 | + | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| O3I04_RS19205 (O3I04_19130) | 15359..15640 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| O3I04_RS19210 (O3I04_19135) | 15650..15943 | + | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| O3I04_RS19215 (O3I04_19140) | 15993..16310 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| O3I04_RS19220 (O3I04_19145) | 16310..18910 | + | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| O3I04_RS19225 (O3I04_19150) | 18928..19641 | + | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| O3I04_RS19230 (O3I04_19155) | 19649..19876 | + | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| O3I04_RS19235 (O3I04_19160) | 19892..20932 | + | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| O3I04_RS19240 (O3I04_19165) | 21015..21161 | + | 147 | WP_001257173 | conjugal transfer protein TraN | - |
| O3I04_RS19245 (O3I04_19170) | 21151..21849 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| O3I04_RS19250 (O3I04_19175) | 21860..22744 | + | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| O3I04_RS19255 (O3I04_19180) | 22744..23904 | + | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| O3I04_RS19260 (O3I04_19185) | 23946..24941 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| O3I04_RS19265 (O3I04_19190) | 24941..25474 | + | 534 | WP_000731969 | phospholipase D family protein | - |
| O3I04_RS19270 (O3I04_19195) | 25667..26212 | + | 546 | WP_001493763 | plasmid pRiA4b ORF-3 family protein | - |
| O3I04_RS19275 (O3I04_19200) | 26349..26921 | + | 573 | WP_001493762 | recombinase family protein | - |
| O3I04_RS19280 (O3I04_19205) | 26958..28349 | + | 1392 | WP_001493761 | ISKra4-like element ISKpn19 family transposase | - |
| O3I04_RS19285 (O3I04_19210) | 28382..28810 | + | 429 | Protein_37 | recombinase family protein | - |
| O3I04_RS19290 (O3I04_19215) | 29129..29785 | - | 657 | WP_001516695 | quinolone resistance pentapeptide repeat protein QnrS1 | - |
Host bacterium
| ID | 1997 | GenBank | NZ_JAPVEN010000034 |
| Plasmid name | L2M4-08|unnamed1 | Incompatibility group | IncN |
| Plasmid size | 52848 bp | Coordinate of oriT [Strand] | 49635..49735 [+] |
| Host baterium | Escherichia coli strain L2M4-08 |
Cargo genes
| Drug resistance gene | qnrS1, blaTEM-1B, tet(A), aph(3'')-Ib, aph(6)-Id |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |