Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101544 |
| Name | oriT1_FWSEC0398|unnamed6 |
| Organism | Escherichia coli strain FWSEC0398 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_RRNV01000100 (22642..22727 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file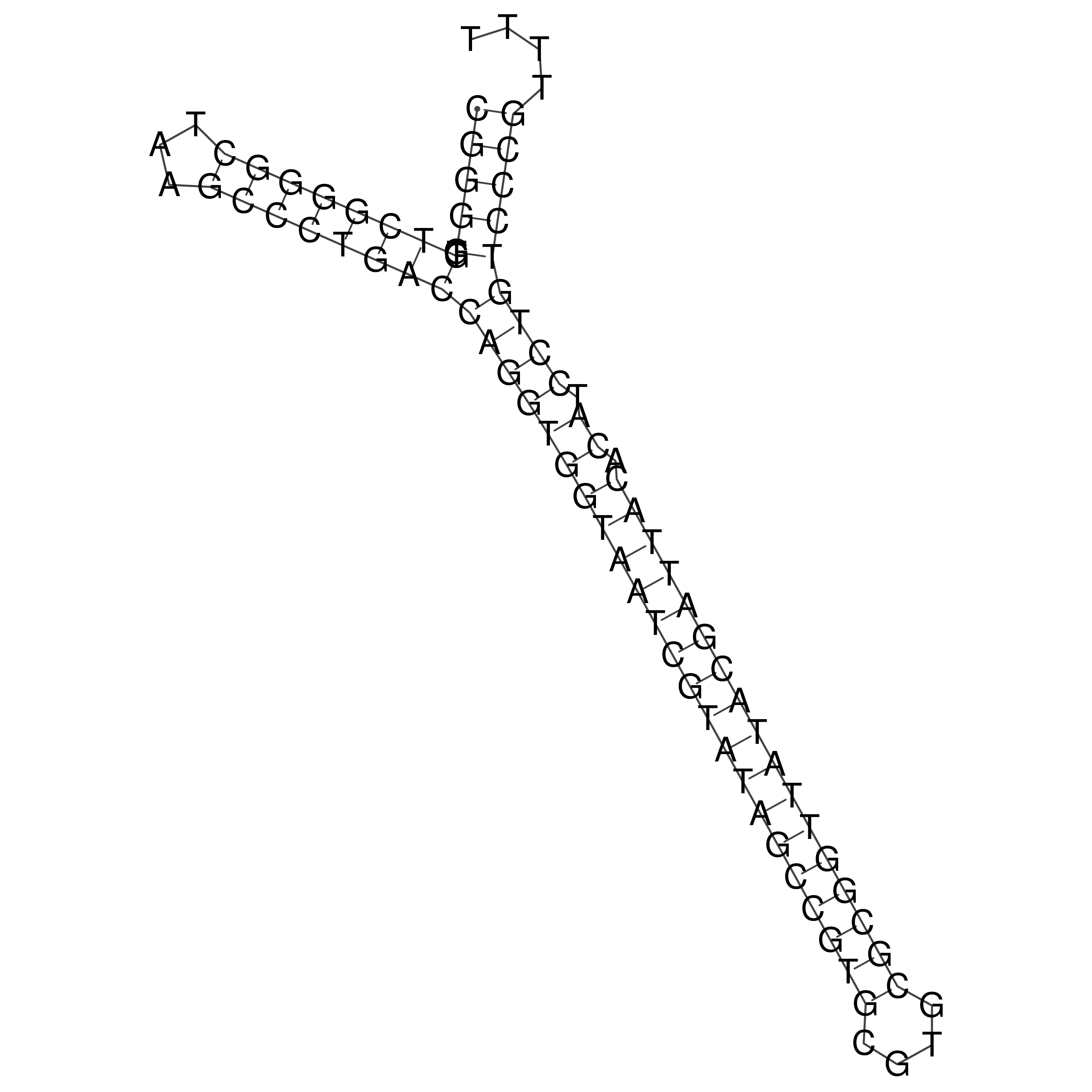
Relaxase
| ID | 1369 | GenBank | WP_136748012 |
| Name | Relaxase_C9173_RS23595_FWSEC0398|unnamed6 |
UniProt ID | _ |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104931.66 Da Isoelectric Point: 8.1134
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEMKAEIPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAAELAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIDQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDKAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRILAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRECREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 483 | GenBank | WP_001291057 |
| Name | WP_001291057_FWSEC0398|unnamed6 |
UniProt ID | A0A142CND4 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12596.69 Da Isoelectric Point: 10.7335
MSEKKTRSGSEKRQKNVLIAVRFSPEEAEIVKEKAEKNGLTVSTLIRKTVLGKQINARIDEDFLKELMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITELANTLRRNLMGR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CND4 |
T4CP
| ID | 997 | GenBank | WP_162534295 |
| Name | t4cp2_C9173_RS24440_FWSEC0398|unnamed6 |
UniProt ID | _ |
| Length | 489 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 489 a.a. Molecular weight: 55147.40 Da Isoelectric Point: 5.5566
MSLQMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQ
FSETFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMILARDLGY
RLEGTDAQPLEVKKYKGSFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSATNTA
TLMQNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLI
RLNAGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPEHVS
SIIGVLTAKPSRKRRKNRTEPYRIIDTFQRRLATAQTSLDLLPQYDTDIESRANELWKKAVHTINNTTRE
ERRVCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSKTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 998 | GenBank | WP_274536677 |
| Name | t4cp1_C9173_RS24515_FWSEC0398|unnamed6 |
UniProt ID | _ |
| Length | 428 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 428 a.a. Molecular weight: 49200.79 Da Isoelectric Point: 10.4408
MSYQHSPTTEDPMNIWYIVGIICLLFALLIWLFLPDIVFATCLILHTMWGMTDWGPFHHFAAPRYNLLAN
TANNAATITFSQWLDVMSQTVRILWLILLPMTIGFLWMWFQHPAQPRFTRRPLNIHTLPHLFSSLSPAIA
PVLADGDSNRLFHGQKRPERRVALTPEVFVAQNNLIRNMQLDVAATRQRFMAQLGQPLTTWKDMAPHEKA
LFAVFGLQFFLGDRKAAVSLMNNLNLSCRLKSKRDQGRFSTPVYSLARNAFIRVIKTEGAQQWLRQHRYR
RINDIGRELWLSMDDLTRHVMFFATTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRF
GREDDIEVINFMNGGKSRSEVIQSGDKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAM
NKALVFGT
Protein domains
No domain identified.
Protein structure
No available structure.
Host bacterium
| ID | 1988 | GenBank | NZ_RRNV01000100 |
| Plasmid name | FWSEC0398|unnamed6 | Incompatibility group | - |
| Plasmid size | 42320 bp | Coordinate of oriT [Strand] | 22642..22727 [-]; 39924..40181 [+] |
| Host baterium | Escherichia coli strain FWSEC0398 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |