Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101483 |
| Name | oriT_pMRVIM0912 |
| Organism | Citrobacter freundii strain MRSN 11938 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JYFY02000005 (66977..67127 [-], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 93..98, 106..111 (TGGCCT..AGGCCA) 12..17, 24..29 (AACCCT..AGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pMRVIM0912
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file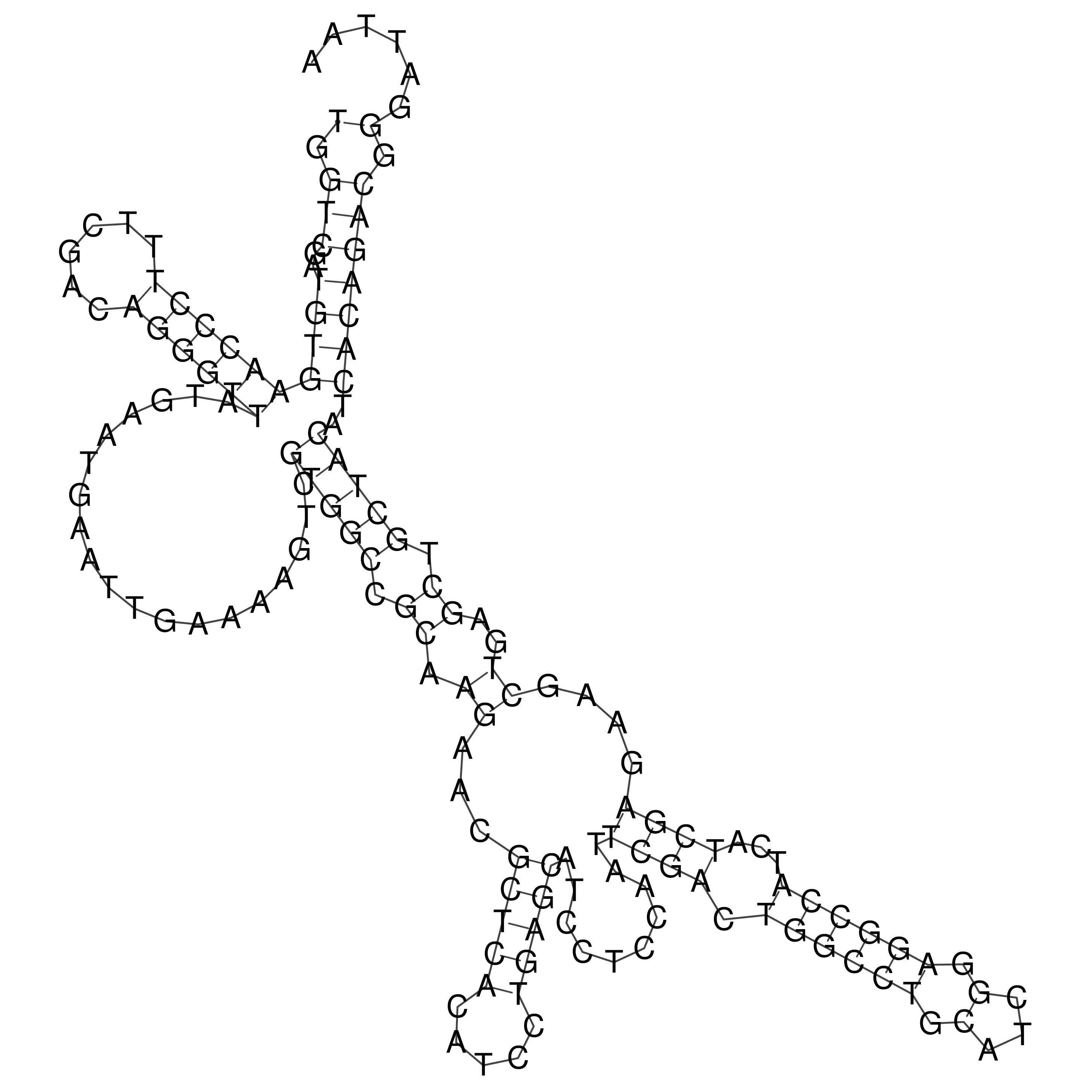
Relaxase
| ID | 1326 | GenBank | WP_001337757 |
| Name | mobH_TN42_RS25940_pMRVIM0912 |
UniProt ID | A0A709FRI2 |
| Length | 992 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 992 a.a. Molecular weight: 109581.85 Da Isoelectric Point: 4.8956
>WP_001337757.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREDPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A709FRI2 |
T4CP
| ID | 944 | GenBank | WP_000637386 |
| Name | traC_TN42_RS25860_pMRVIM0912 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92590.87 Da Isoelectric Point: 5.9222
>WP_000637386.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTERIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 945 | GenBank | WP_000178856 |
| Name | traD_TN42_RS25935_pMRVIM0912 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69197.66 Da Isoelectric Point: 6.9403
>WP_000178856.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 43823..69043
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| TN42_RS25800 (TN42_24150) | 38950..39858 | - | 909 | WP_000739139 | hypothetical protein | - |
| TN42_RS25805 (TN42_24155) | 39869..40837 | - | 969 | WP_000085162 | AAA family ATPase | - |
| TN42_RS25810 (TN42_24160) | 41057..41242 | - | 186 | WP_001186917 | hypothetical protein | - |
| TN42_RS25815 (TN42_24165) | 41546..41866 | - | 321 | WP_000547566 | hypothetical protein | - |
| TN42_RS25820 (TN42_24170) | 42160..42801 | + | 642 | WP_000796664 | hypothetical protein | - |
| TN42_RS25825 (TN42_24175) | 42924..43784 | + | 861 | WP_000709517 | hypothetical protein | - |
| TN42_RS25830 (TN42_24180) | 43823..46621 | - | 2799 | WP_001256487 | conjugal transfer mating pair stabilization protein TraN | traN |
| TN42_RS31720 | 46737..46979 | - | 243 | WP_046155150 | TraU family protein | traU |
| TN42_RS25835 (TN42_19725) | 47005..47770 | - | 766 | WP_044702044 | TraU family protein | traU |
| TN42_RS25840 (TN42_19730) | 47770..48441 | - | 672 | WP_001337754 | EAL domain-containing protein | - |
| TN42_RS25845 (TN42_19735) | 48438..49703 | - | 1266 | WP_000621288 | TrbC family F-type conjugative pilus assembly protein | traW |
| TN42_RS25850 (TN42_19740) | 49666..50196 | - | 531 | WP_001010738 | S26 family signal peptidase | - |
| TN42_RS25855 (TN42_19745) | 50193..50510 | - | 318 | WP_000351984 | hypothetical protein | - |
| TN42_RS25860 (TN42_19750) | 50525..52972 | - | 2448 | WP_000637386 | type IV secretion system protein TraC | virb4 |
| TN42_RS25865 (TN42_19755) | 52969..53676 | - | 708 | WP_001259347 | DsbC family protein | - |
| TN42_RS25870 (TN42_19760) | 53825..59356 | - | 5532 | WP_000606833 | Ig-like domain-containing protein | - |
| TN42_RS25875 (TN42_19765) | 59557..59949 | - | 393 | WP_000479535 | TraA family conjugative transfer protein | - |
| TN42_RS25880 (TN42_19770) | 59953..60531 | - | 579 | WP_000793435 | type IV conjugative transfer system lipoprotein TraV | traV |
| TN42_RS25885 (TN42_19775) | 60528..61841 | - | 1314 | WP_024131605 | TraB/VirB10 family protein | traB |
| TN42_RS25890 (TN42_19780) | 61841..62758 | - | 918 | WP_000794249 | type-F conjugative transfer system secretin TraK | traK |
| TN42_RS25895 (TN42_19785) | 62742..63368 | - | 627 | WP_001049716 | TraE/TraK family type IV conjugative transfer system protein | traE |
| TN42_RS25900 (TN42_19790) | 63365..63646 | - | 282 | WP_000805625 | type IV conjugative transfer system protein TraL | traL |
| TN42_RS25905 (TN42_19795) | 63791..64156 | - | 366 | WP_001052531 | hypothetical protein | - |
| TN42_RS31105 | 64168..64311 | - | 144 | WP_001275801 | hypothetical protein | - |
| TN42_RS25910 (TN42_19800) | 64492..65154 | - | 663 | WP_001231463 | hypothetical protein | - |
| TN42_RS25915 (TN42_19805) | 65154..65531 | - | 378 | WP_000869261 | hypothetical protein | - |
| TN42_RS25920 (TN42_19810) | 65541..65987 | - | 447 | WP_000122506 | hypothetical protein | - |
| TN42_RS25925 (TN42_19815) | 65997..66626 | - | 630 | WP_000743450 | DUF4400 domain-containing protein | tfc7 |
| TN42_RS25930 (TN42_19820) | 66583..67128 | - | 546 | WP_000228720 | hypothetical protein | - |
| TN42_RS25935 (TN42_19825) | 67178..69043 | - | 1866 | WP_000178856 | conjugative transfer system coupling protein TraD | virb4 |
| TN42_RS25940 (TN42_19830) | 69040..72018 | - | 2979 | WP_001337757 | MobH family relaxase | - |
| TN42_RS25945 (TN42_19835) | 72186..72803 | + | 618 | WP_001249396 | hypothetical protein | - |
| TN42_RS25950 (TN42_19840) | 72785..73018 | + | 234 | WP_001191892 | hypothetical protein | - |
Host bacterium
| ID | 1927 | GenBank | NZ_JYFY02000005 |
| Plasmid name | pMRVIM0912 | Incompatibility group | IncA/C2 |
| Plasmid size | 151387 bp | Coordinate of oriT [Strand] | 66977..67127 [-] |
| Host baterium | Citrobacter freundii strain MRSN 11938 |
Cargo genes
| Drug resistance gene | catA1, tet(C), blaCTX-M-39 |
| Virulence gene | - |
| Metal resistance gene | merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |