Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101427 |
| Name | oriT_pCRE19_1 |
| Organism | Klebsiella pneumoniae strain S19_CRE19 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAHBNP010000002 (140702..140751 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pCRE19_1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file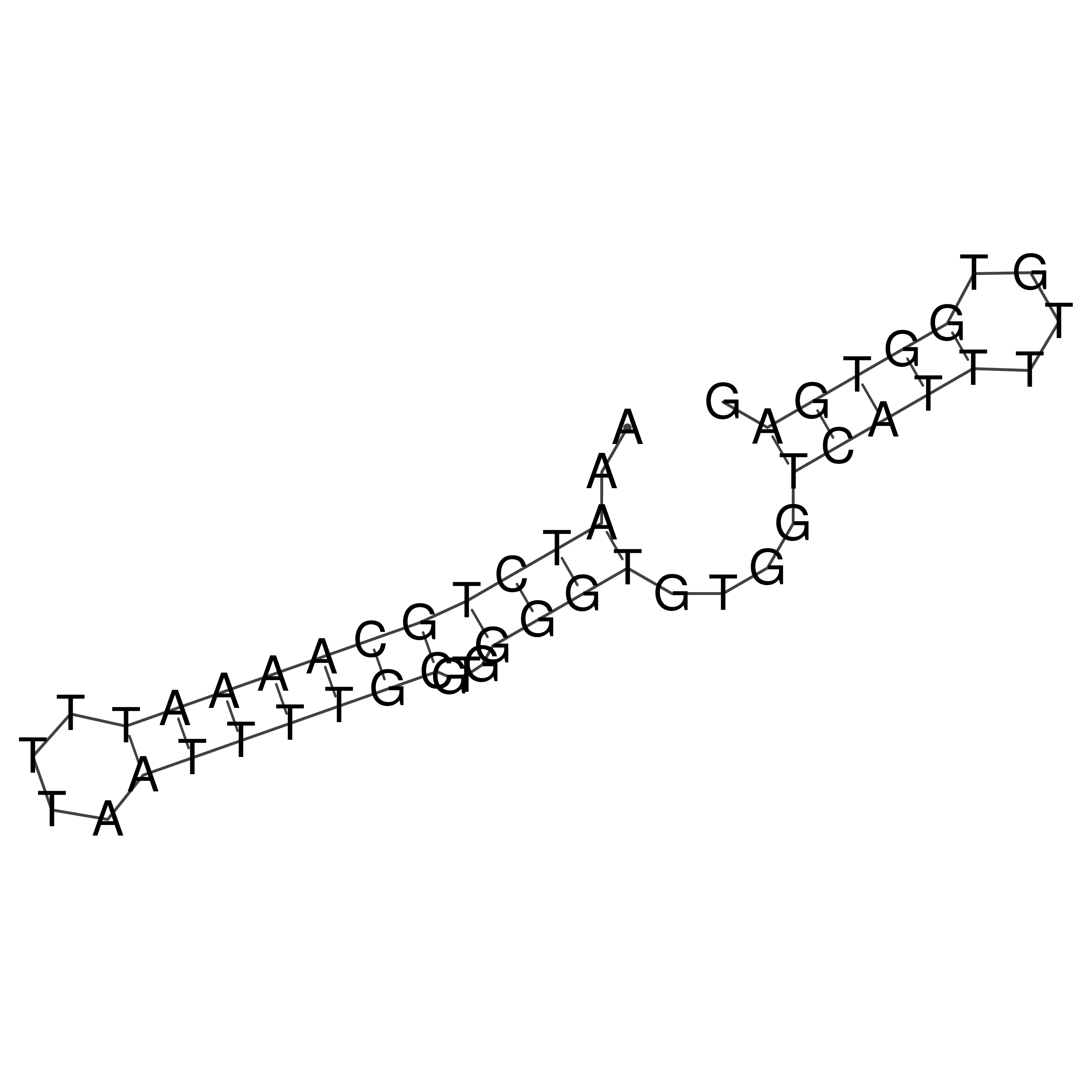
Relaxase
| ID | 1295 | GenBank | WP_004153029 |
| Name | traI_KIN62_RS27790_pCRE19_1 |
UniProt ID | A0A422WEA5 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190808.90 Da Isoelectric Point: 6.2081
>WP_004153029.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 909 | GenBank | WP_004152630 |
| Name | traD_KIN62_RS27785_pCRE19_1 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85951.02 Da Isoelectric Point: 5.0632
>WP_004152630.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEEREAEGSLARALFTPDAPASGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAAEPSVRATEPPVLRVTTVPLIKPKAAAAAAAASTASSAGIPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEEREAEGSLARALFTPDAPASGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAAEPSVRATEPPVLRVTTVPLIKPKAAAAAAAASTASSAGIPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 140144..168562
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KIN62_RS27575 (KIN62_27505) | 135161..135511 | + | 351 | WP_004152758 | hypothetical protein | - |
| KIN62_RS27580 (KIN62_27510) | 136147..136503 | + | 357 | WP_004152717 | hypothetical protein | - |
| KIN62_RS27585 (KIN62_27515) | 136564..136776 | + | 213 | WP_004152718 | hypothetical protein | - |
| KIN62_RS27590 (KIN62_27520) | 136787..137011 | + | 225 | WP_004152719 | hypothetical protein | - |
| KIN62_RS27595 (KIN62_27525) | 137092..137412 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| KIN62_RS27600 (KIN62_27530) | 137402..137680 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| KIN62_RS27605 (KIN62_27535) | 137681..138094 | + | 414 | WP_004152722 | helix-turn-helix domain-containing protein | - |
| KIN62_RS27610 (KIN62_27540) | 138928..139749 | + | 822 | WP_004178064 | DUF932 domain-containing protein | - |
| KIN62_RS27615 (KIN62_27545) | 139782..140111 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| KIN62_RS27620 (KIN62_27550) | 140144..140629 | - | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| KIN62_RS27625 (KIN62_27555) | 141063..141455 | + | 393 | WP_004178062 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KIN62_RS27630 (KIN62_27560) | 141669..142364 | + | 696 | WP_004178061 | transcriptional regulator TraJ family protein | - |
| KIN62_RS27635 (KIN62_27565) | 142448..142819 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| KIN62_RS27640 (KIN62_27570) | 142873..143241 | + | 369 | WP_004178060 | type IV conjugative transfer system pilin TraA | - |
| KIN62_RS27645 (KIN62_27575) | 143255..143560 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| KIN62_RS27650 (KIN62_27580) | 143580..144146 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| KIN62_RS27655 (KIN62_27585) | 144133..144873 | + | 741 | WP_004152601 | type-F conjugative transfer system secretin TraK | traK |
| KIN62_RS27660 (KIN62_27590) | 144873..146297 | + | 1425 | WP_004152600 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KIN62_RS27665 (KIN62_27595) | 146290..146886 | + | 597 | WP_004152599 | conjugal transfer pilus-stabilizing protein TraP | - |
| KIN62_RS27670 (KIN62_27600) | 146909..147478 | + | 570 | WP_004152598 | type IV conjugative transfer system lipoprotein TraV | traV |
| KIN62_RS27675 (KIN62_27605) | 147610..148020 | + | 411 | WP_004152597 | lipase chaperone | - |
| KIN62_RS27680 (KIN62_27610) | 148025..148315 | + | 291 | WP_004152596 | hypothetical protein | - |
| KIN62_RS27685 (KIN62_27615) | 148339..148557 | + | 219 | WP_004171484 | hypothetical protein | - |
| KIN62_RS27690 (KIN62_27620) | 148558..148875 | + | 318 | WP_004152595 | hypothetical protein | - |
| KIN62_RS27695 (KIN62_27625) | 148942..149346 | + | 405 | WP_004152594 | hypothetical protein | - |
| KIN62_RS27700 (KIN62_27630) | 149642..150040 | + | 399 | WP_004153071 | hypothetical protein | - |
| KIN62_RS27705 (KIN62_27635) | 150112..152751 | + | 2640 | WP_004152592 | type IV secretion system protein TraC | virb4 |
| KIN62_RS27710 (KIN62_27640) | 152751..153140 | + | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| KIN62_RS27715 (KIN62_27645) | 153140..153766 | + | 627 | WP_004152590 | type-F conjugative transfer system protein TraW | traW |
| KIN62_RS27720 (KIN62_27650) | 153810..154769 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| KIN62_RS27725 (KIN62_27655) | 154782..155420 | + | 639 | WP_004152682 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KIN62_RS27730 (KIN62_27660) | 155468..157423 | + | 1956 | WP_004153093 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| KIN62_RS27735 (KIN62_27665) | 157455..157691 | + | 237 | WP_004152683 | conjugal transfer protein TrbE | - |
| KIN62_RS29205 | 157688..157873 | + | 186 | WP_004152684 | hypothetical protein | - |
| KIN62_RS27740 (KIN62_27670) | 157919..158245 | + | 327 | WP_004152685 | hypothetical protein | - |
| KIN62_RS27745 (KIN62_27675) | 158266..159018 | + | 753 | WP_004152686 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KIN62_RS27750 (KIN62_27680) | 159029..159268 | + | 240 | WP_004152687 | type-F conjugative transfer system pilin chaperone TraQ | - |
| KIN62_RS27755 (KIN62_27685) | 159240..159812 | + | 573 | WP_004152688 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| KIN62_RS27760 (KIN62_27690) | 159805..160233 | + | 429 | WP_004152689 | hypothetical protein | - |
| KIN62_RS27765 (KIN62_27695) | 160220..161590 | + | 1371 | WP_004178055 | conjugal transfer pilus assembly protein TraH | traH |
| KIN62_RS27770 (KIN62_27700) | 161590..164439 | + | 2850 | WP_004178054 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KIN62_RS27775 (KIN62_27705) | 164445..164972 | + | 528 | WP_004153030 | hypothetical protein | - |
| KIN62_RS27780 (KIN62_27710) | 165162..165893 | + | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| KIN62_RS27785 (KIN62_27715) | 166253..168562 | + | 2310 | WP_004152630 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 1871 | GenBank | NZ_JAHBNP010000002 |
| Plasmid name | pCRE19_1 | Incompatibility group | IncFIB |
| Plasmid size | 211957 bp | Coordinate of oriT [Strand] | 140702..140751 [-] |
| Host baterium | Klebsiella pneumoniae strain S19_CRE19 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsR, arsD, arsA, arsB, arsC, silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsH, fecE, fecD |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |