Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101307 |
| Name | oriT_SCPM-O-B-8045|unnamed |
| Organism | Klebsiella pneumoniae strain SCPM-O-B-8045 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_NPII01000022 (26947..26996 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_SCPM-O-B-8045|unnamed
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file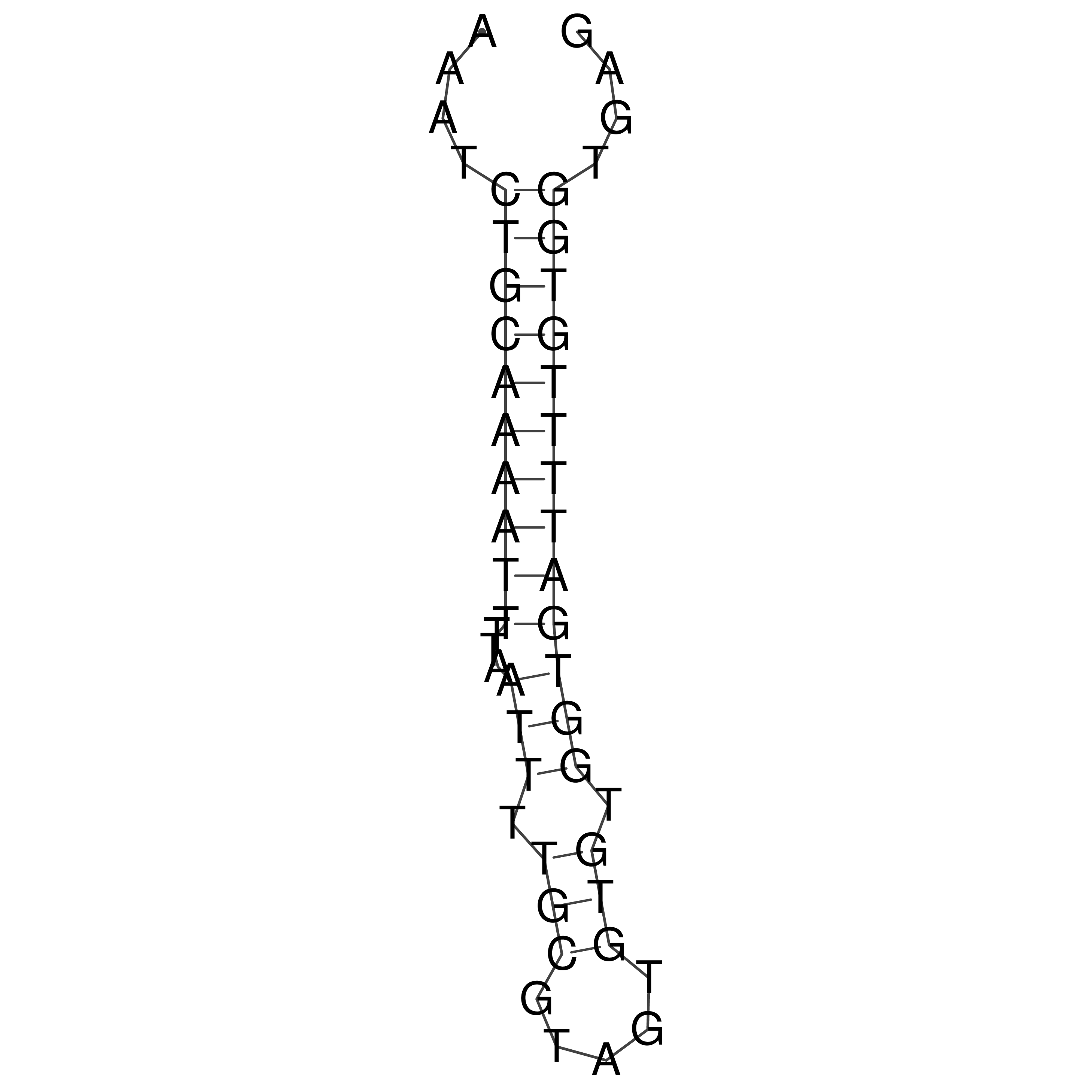
Relaxase
| ID | 1241 | GenBank | WP_065519309 |
| Name | traI_CIT28_RS24455_SCPM-O-B-8045|unnamed |
UniProt ID | A0A809SXI2 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190492.21 Da Isoelectric Point: 6.0263
>WP_065519309.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 852 | GenBank | WP_013214038 |
| Name | traD_CIT28_RS24450_SCPM-O-B-8045|unnamed |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86010.07 Da Isoelectric Point: 5.1787
>WP_013214038.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26389..55001
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CIT28_RS24215 (CIT28_24195) | 21545..21862 | + | 318 | WP_014343505 | hypothetical protein | - |
| CIT28_RS24220 (CIT28_24200) | 22703..23059 | + | 357 | WP_014343501 | hypothetical protein | - |
| CIT28_RS24225 (CIT28_24205) | 23120..23332 | + | 213 | WP_014343500 | hypothetical protein | - |
| CIT28_RS24230 (CIT28_24210) | 23343..23567 | + | 225 | WP_014343499 | hypothetical protein | - |
| CIT28_RS24235 (CIT28_24215) | 23648..23968 | + | 321 | WP_014343498 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| CIT28_RS24240 (CIT28_24220) | 23958..24236 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| CIT28_RS30360 (CIT28_24225) | 24240..24344 | + | 105 | Protein_37 | transcriptional regulator | - |
| CIT28_RS24265 (CIT28_24245) | 25173..25994 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| CIT28_RS24270 (CIT28_24250) | 26027..26356 | + | 330 | WP_013214015 | DUF5983 family protein | - |
| CIT28_RS24275 (CIT28_24255) | 26389..26874 | - | 486 | WP_014343495 | transglycosylase SLT domain-containing protein | virB1 |
| CIT28_RS24280 (CIT28_24260) | 27265..27681 | + | 417 | WP_014343494 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| CIT28_RS24285 (CIT28_24265) | 27881..28618 | + | 738 | WP_013214018 | conjugal transfer protein TrbJ | - |
| CIT28_RS24290 (CIT28_24270) | 28733..28939 | + | 207 | WP_171773970 | TraY domain-containing protein | - |
| CIT28_RS24295 (CIT28_24275) | 29001..29369 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| CIT28_RS24300 (CIT28_24280) | 29383..29688 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| CIT28_RS24305 (CIT28_24285) | 29708..30274 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| CIT28_RS24310 (CIT28_24290) | 30261..31001 | + | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| CIT28_RS24315 (CIT28_24295) | 31001..32425 | + | 1425 | WP_013214020 | F-type conjugal transfer pilus assembly protein TraB | traB |
| CIT28_RS24320 (CIT28_24300) | 32497..33081 | + | 585 | WP_014343493 | type IV conjugative transfer system lipoprotein TraV | traV |
| CIT28_RS24325 (CIT28_24305) | 33404..33622 | + | 219 | WP_004195468 | hypothetical protein | - |
| CIT28_RS24330 (CIT28_24310) | 33623..33934 | + | 312 | WP_013214022 | hypothetical protein | - |
| CIT28_RS24335 (CIT28_24315) | 34001..34405 | + | 405 | WP_004197817 | hypothetical protein | - |
| CIT28_RS24350 (CIT28_24330) | 34782..35180 | + | 399 | WP_014343490 | hypothetical protein | - |
| CIT28_RS24355 (CIT28_24335) | 35252..37891 | + | 2640 | WP_013214024 | type IV secretion system protein TraC | virb4 |
| CIT28_RS24360 (CIT28_24340) | 37891..38280 | + | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| CIT28_RS24365 (CIT28_24345) | 38280..38915 | + | 636 | WP_013214026 | type-F conjugative transfer system protein TraW | traW |
| CIT28_RS24370 (CIT28_24350) | 38950..39351 | + | 402 | WP_004194979 | hypothetical protein | - |
| CIT28_RS24375 (CIT28_24355) | 39348..40337 | + | 990 | WP_013214027 | conjugal transfer pilus assembly protein TraU | traU |
| CIT28_RS24380 (CIT28_24360) | 40350..40988 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| CIT28_RS24385 (CIT28_24365) | 41047..43002 | + | 1956 | WP_014343489 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| CIT28_RS24390 (CIT28_24370) | 43034..43288 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| CIT28_RS24395 (CIT28_24375) | 43266..43514 | + | 249 | WP_013214029 | hypothetical protein | - |
| CIT28_RS24400 (CIT28_24380) | 43527..43853 | + | 327 | WP_013214030 | hypothetical protein | - |
| CIT28_RS24405 (CIT28_24385) | 43874..44626 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| CIT28_RS24410 (CIT28_24390) | 44637..44876 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| CIT28_RS24415 (CIT28_24395) | 44848..45405 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| CIT28_RS24420 (CIT28_24400) | 45451..45894 | + | 444 | WP_013214032 | F-type conjugal transfer protein TrbF | - |
| CIT28_RS24425 (CIT28_24405) | 45872..47251 | + | 1380 | WP_074160414 | conjugal transfer pilus assembly protein TraH | traH |
| CIT28_RS24430 (CIT28_24410) | 47251..50073 | + | 2823 | WP_013214034 | conjugal transfer mating-pair stabilization protein TraG | traG |
| CIT28_RS24435 (CIT28_24415) | 50096..50698 | + | 603 | WP_013214035 | conjugal transfer protein TraS | - |
| CIT28_RS24440 (CIT28_24420) | 50949..51680 | + | 732 | WP_013214036 | conjugal transfer complement resistance protein TraT | - |
| CIT28_RS29990 | 51873..52562 | + | 690 | WP_014343485 | hypothetical protein | - |
| CIT28_RS24450 (CIT28_24430) | 52689..55001 | + | 2313 | WP_013214038 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 1751 | GenBank | NZ_NPII01000022 |
| Plasmid name | SCPM-O-B-8045|unnamed | Incompatibility group | IncFIB |
| Plasmid size | 82279 bp | Coordinate of oriT [Strand] | 26947..26996 [-] |
| Host baterium | Klebsiella pneumoniae strain SCPM-O-B-8045 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |