Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101265 |
| Name | oriT_PX02|unnamed1 |
| Organism | Raoultella ornithinolytica strain PX02 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_NJBC01000001 (1931..1980 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_PX02|unnamed1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file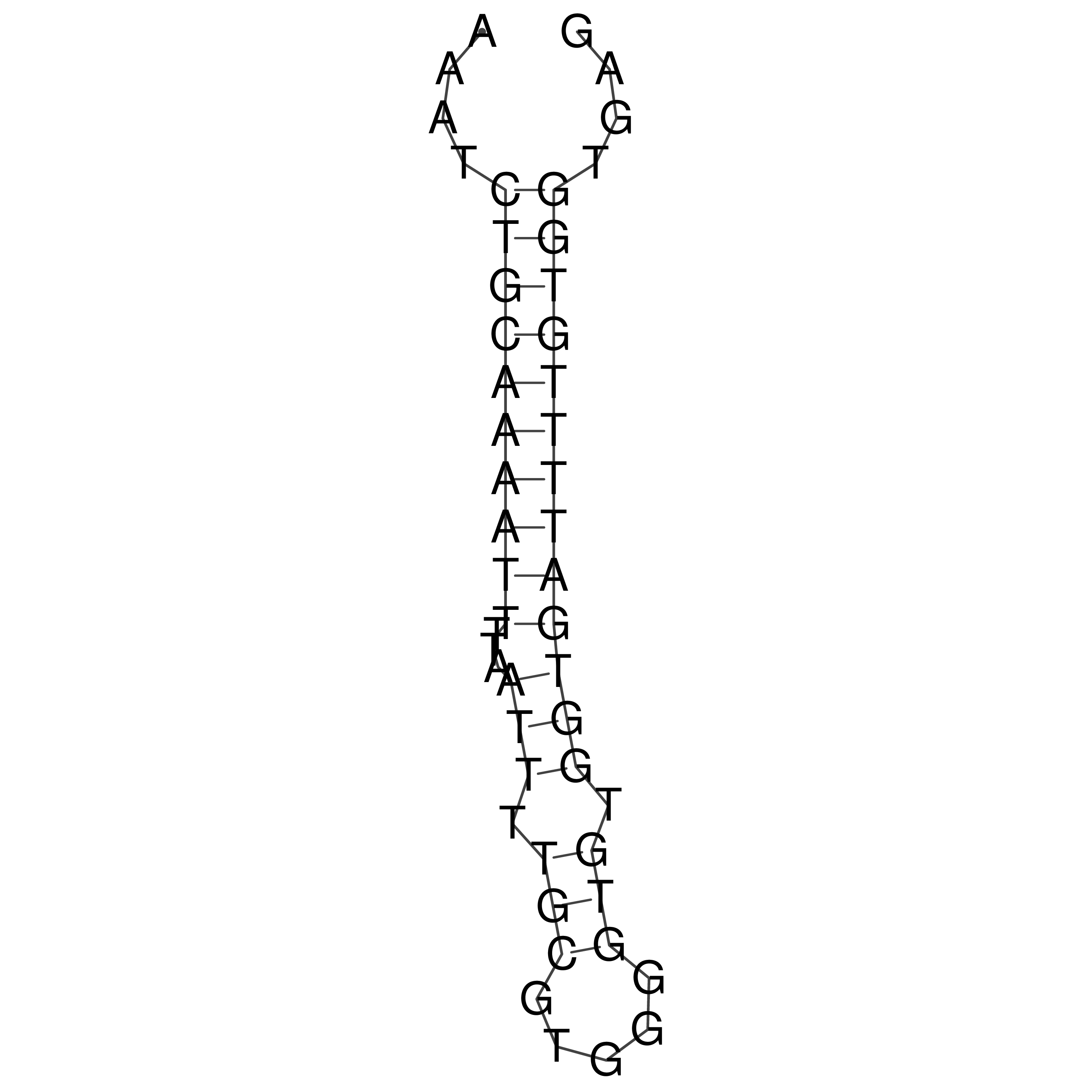
Relaxase
| ID | 1226 | GenBank | WP_088401876 |
| Name | traI_CEG93_RS00180_PX02|unnamed1 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190588.30 Da Isoelectric Point: 5.9980
>WP_088401876.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella/Raoultella group]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQSSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTIK
KQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIRK
ALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHDARRENGETGREEITLP
VLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAITA
HGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQSSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLHTPAEQAIHLSIPLLESQDLTFTRPQLLATALETGGDKVPMWSIEMTIK
KQIKSGQLLNVGVSPGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILETPDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPHVIGLGPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSISPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDIDRALATIEQVTPERVPRKEGAWVPGSSVVEFTPTQEEEIRK
ALSKGESLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNNDRRALNGLIHDARRENGETGREEITLP
VLVTSNIRDGELRKLSTWTAHKGAVALVDNVYHRIDNVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEEITVSQGDRMRFSKSDPERGYVANSVWEVQSVSGDSVTLSDGKTTRTLTPKADEAQQHIDLAYAITA
HGAQGASEPYAIALEGMVGARKLMASFESAYVGLSRMKQHVQVYTDRREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNIGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASETLQRDRLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 836 | GenBank | WP_088401875 |
| Name | traD_CEG93_RS00175_PX02|unnamed1 |
UniProt ID | _ |
| Length | 760 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 760 a.a. Molecular weight: 85026.96 Da Isoelectric Point: 5.0080
>WP_088401875.1 type IV conjugative transfer system coupling protein TraD [Raoultella ornithinolytica]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFATGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPEPGPADTESHASEQPESVSPPA
PAEMTVTPAPVKAPPTTKRPAAEPPVLPVTPVPLLNQKAAAAAATASSAGTPAAAAGGTEQELAQQPTEQ
GQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDIEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFATGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPEPGPADTESHASEQPESVSPPA
PAEMTVTPAPVKAPPTTKRPAAEPPVLPVTPVPLLNQKAAAAAATASSAGTPAAAAGGTEQELAQQPTEQ
GQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDIEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1373..29033
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CEG93_RS00005 (CEG93_00005) | 157..978 | + | 822 | WP_015065622 | DUF932 domain-containing protein | - |
| CEG93_RS30020 | 1026..1340 | + | 315 | WP_001568110 | DUF5983 family protein | - |
| CEG93_RS00010 (CEG93_00010) | 1373..1858 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| CEG93_RS00015 (CEG93_00015) | 2249..2665 | + | 417 | WP_074181767 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| CEG93_RS00020 (CEG93_00020) | 2865..3584 | + | 720 | WP_172412520 | conjugal transfer protein TrbJ | - |
| CEG93_RS00025 (CEG93_00025) | 3717..3923 | + | 207 | WP_172412524 | TraY domain-containing protein | - |
| CEG93_RS00030 (CEG93_00030) | 3985..4353 | + | 369 | WP_004178060 | type IV conjugative transfer system pilin TraA | - |
| CEG93_RS00035 (CEG93_00035) | 4367..4672 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| CEG93_RS00040 (CEG93_00040) | 4692..5258 | + | 567 | WP_004194424 | type IV conjugative transfer system protein TraE | traE |
| CEG93_RS00045 (CEG93_00045) | 5245..5985 | + | 741 | WP_088401865 | type-F conjugative transfer system secretin TraK | traK |
| CEG93_RS00050 (CEG93_00050) | 5985..7409 | + | 1425 | WP_088401866 | F-type conjugal transfer pilus assembly protein TraB | traB |
| CEG93_RS00055 (CEG93_00055) | 7482..8051 | + | 570 | WP_032456782 | type IV conjugative transfer system lipoprotein TraV | traV |
| CEG93_RS30735 | 8206..8637 | + | 432 | WP_233435409 | hypothetical protein | - |
| CEG93_RS00065 (CEG93_00065) | 8661..8879 | + | 219 | WP_032456783 | hypothetical protein | - |
| CEG93_RS00070 (CEG93_00070) | 8880..9191 | + | 312 | WP_088401867 | hypothetical protein | - |
| CEG93_RS00075 (CEG93_00075) | 9258..9662 | + | 405 | WP_032456785 | hypothetical protein | - |
| CEG93_RS00080 (CEG93_00080) | 9705..10004 | + | 300 | WP_088401868 | hypothetical protein | - |
| CEG93_RS00085 (CEG93_00085) | 10012..10410 | + | 399 | WP_172412521 | hypothetical protein | - |
| CEG93_RS00090 (CEG93_00090) | 10482..13121 | + | 2640 | WP_088401870 | type IV secretion system protein TraC | virb4 |
| CEG93_RS00095 (CEG93_00095) | 13121..13510 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| CEG93_RS00100 (CEG93_00100) | 13510..14136 | + | 627 | WP_015065633 | type-F conjugative transfer system protein TraW | traW |
| CEG93_RS00105 (CEG93_00105) | 14180..15139 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| CEG93_RS00110 (CEG93_00110) | 15152..15790 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| CEG93_RS00115 (CEG93_00115) | 15849..17804 | + | 1956 | WP_088401871 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| CEG93_RS00120 (CEG93_00120) | 17836..18072 | + | 237 | WP_016529263 | conjugal transfer protein TrbE | - |
| CEG93_RS31105 (CEG93_00125) | 18069..18254 | + | 186 | WP_086070806 | hypothetical protein | - |
| CEG93_RS00130 (CEG93_00130) | 18300..18626 | + | 327 | WP_016529362 | hypothetical protein | - |
| CEG93_RS00135 (CEG93_00135) | 18647..19399 | + | 753 | WP_088401872 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| CEG93_RS00140 (CEG93_00140) | 19410..19649 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| CEG93_RS00145 (CEG93_00145) | 19621..20193 | + | 573 | WP_032408763 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| CEG93_RS00150 (CEG93_00150) | 20186..20614 | + | 429 | WP_004152689 | hypothetical protein | - |
| CEG93_RS00155 (CEG93_00155) | 20601..21971 | + | 1371 | WP_004178055 | conjugal transfer pilus assembly protein TraH | traH |
| CEG93_RS00160 (CEG93_00160) | 21971..24814 | + | 2844 | WP_088401873 | conjugal transfer mating-pair stabilization protein TraG | traG |
| CEG93_RS00165 (CEG93_00165) | 24825..25349 | + | 525 | WP_015632518 | conjugal transfer entry exclusion protein | - |
| CEG93_RS00170 (CEG93_00170) | 25658..26389 | + | 732 | WP_088401874 | conjugal transfer complement resistance protein TraT | - |
| CEG93_RS00175 (CEG93_00175) | 26751..29033 | + | 2283 | WP_088401875 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 1709 | GenBank | NZ_NJBC01000001 |
| Plasmid name | PX02|unnamed1 | Incompatibility group | IncFII |
| Plasmid size | 75035 bp | Coordinate of oriT [Strand] | 1931..1980 [-] |
| Host baterium | Raoultella ornithinolytica strain PX02 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |