Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101242 |
| Name | oriT_pTR3 |
| Organism | Klebsiella pneumoniae JT4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAPZ01000355 (4180..4262 [-], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | 27..34, 37..44 (AGTGCGAT..ATCGCACT) 13..18, 32..37 (TAAATC..GATTTA) |
| Location of nic site | 55..56 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 83 nt
>oriT_pTR3
TTTTTTGTTTTTTAAATCAGTTAGTTAGTGCGATTTATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
TTTTTTGTTTTTTAAATCAGTTAGTTAGTGCGATTTATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file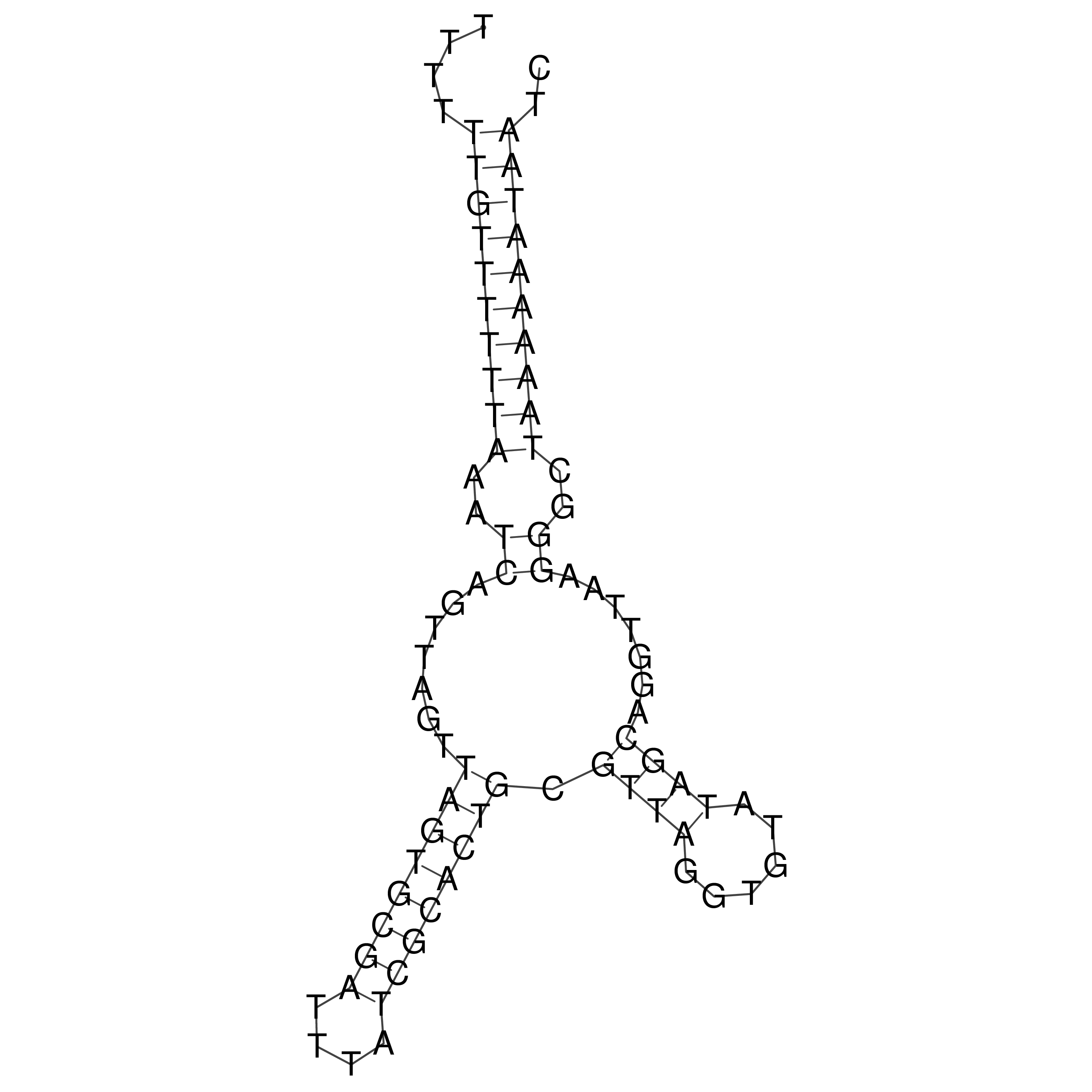
Relaxase
| ID | 1214 | GenBank | WP_014014966 |
| Name | mobF_AH61_RS30290_pTR3 |
UniProt ID | I7CZU5 |
| Length | 1068 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1068 a.a. Molecular weight: 118452.14 Da Isoelectric Point: 6.4309
>WP_014014966.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGEIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDIEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNNDQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTAQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGFNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFLGDTSQTKAVEAGKPFEQLISAGMQTSYMKDIQ
RQKNDVLLEAVKLAAEGHISGSLARLSNISAEKDQDKRLEAVANRWLSFSPEQREGTLIISGTNESRVIL
NSQIREALQLQGKGVEVNFLERVDSTQAERRDSKYYQVGQIIIPEKDYKNGLQRGESYRVLDTGPGNLLT
VAGSDGQKISFSPRTHKNLSVYTSVAAELAVGDKVMVTRNDKTLDVANGDRFTVTGTTEKGTFTLTNAKG
REIELDQKQAAYLSYAYATTVHKAQGLTCDRVLFNIDTRSLTTSKDVFYVGISRARHEVEIFTDDKKRLL
SSASRNSPKTTASEIDRFLGMEERYKDVNRSDGQQEKSYGHDAGHQTEEKENTMKNATEAAHDGPDTSGT
ENTSQDLRQREQEEVNAAIRYEQRQEATPEMEANPYHDYERYADEGPDYGDMQDYSTYEEYERARDVELP
QHEHENHQHDHDKEGYGL
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGEIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDIEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNNDQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTAQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGFNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFLGDTSQTKAVEAGKPFEQLISAGMQTSYMKDIQ
RQKNDVLLEAVKLAAEGHISGSLARLSNISAEKDQDKRLEAVANRWLSFSPEQREGTLIISGTNESRVIL
NSQIREALQLQGKGVEVNFLERVDSTQAERRDSKYYQVGQIIIPEKDYKNGLQRGESYRVLDTGPGNLLT
VAGSDGQKISFSPRTHKNLSVYTSVAAELAVGDKVMVTRNDKTLDVANGDRFTVTGTTEKGTFTLTNAKG
REIELDQKQAAYLSYAYATTVHKAQGLTCDRVLFNIDTRSLTTSKDVFYVGISRARHEVEIFTDDKKRLL
SSASRNSPKTTASEIDRFLGMEERYKDVNRSDGQQEKSYGHDAGHQTEEKENTMKNATEAAHDGPDTSGT
ENTSQDLRQREQEEVNAAIRYEQRQEATPEMEANPYHDYERYADEGPDYGDMQDYSTYEEYERARDVELP
QHEHENHQHDHDKEGYGL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | I7CZU5 |
T4CP
| ID | 825 | GenBank | WP_014014965 |
| Name | t4cp2_AH61_RS30285_pTR3 |
UniProt ID | _ |
| Length | 510 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 510 a.a. Molecular weight: 58174.11 Da Isoelectric Point: 9.7081
>WP_014014965.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MTEQERGLIFLCSLLGPIPFVWFITAKFVYGINSETAHYLVPYLLKNTFNLWPLWVAIIAGAVIGLCGFI
GFIIYDKGRVYKGPRFKRVLRGTKLVRASSLAERTKERGHKQLTVAGVPVPTEYETLHFSIGGATGTGKS
TIIKEAIYSSLKRGRDKRVILDPNGEYVETFYRENDIILNPYDKRSPGWSMFNEIRDEFDYDLYAVSIIQ
ESKERSAEEWYRYGRLLYREVSKKLLTHKRNPSMEEVLYWCNVAEYDKVKDFVKGTPAESLFIKGAERAT
GSARFVLGDKLASHLKMPEGNFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTFSQLVERYGINIAETLLGSMRSGIVMGSSRL
GKATLEHMSRALGEIEGEIERRQGGILQSGPNNRPRIETRTVRAVTPTEISELRNLTGYIYFPGDLPVGK
FKTKHVQYTRKNPVPGIIMR
MTEQERGLIFLCSLLGPIPFVWFITAKFVYGINSETAHYLVPYLLKNTFNLWPLWVAIIAGAVIGLCGFI
GFIIYDKGRVYKGPRFKRVLRGTKLVRASSLAERTKERGHKQLTVAGVPVPTEYETLHFSIGGATGTGKS
TIIKEAIYSSLKRGRDKRVILDPNGEYVETFYRENDIILNPYDKRSPGWSMFNEIRDEFDYDLYAVSIIQ
ESKERSAEEWYRYGRLLYREVSKKLLTHKRNPSMEEVLYWCNVAEYDKVKDFVKGTPAESLFIKGAERAT
GSARFVLGDKLASHLKMPEGNFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTFSQLVERYGINIAETLLGSMRSGIVMGSSRL
GKATLEHMSRALGEIEGEIERRQGGILQSGPNNRPRIETRTVRAVTPTEISELRNLTGYIYFPGDLPVGK
FKTKHVQYTRKNPVPGIIMR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 20477..30759
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AH61_RS30340 | 15830..16642 | - | 813 | WP_004201164 | subclass B1 metallo-beta-lactamase NDM-1 | - |
| AH61_RS30350 | 16938..17969 | + | 1032 | WP_001395480 | IS630-like element ISEc33 family transposase | - |
| AH61_RS32445 | 17988..18734 | - | 747 | Protein_18 | IS30-like element ISAba125 family transposase | - |
| AH61_RS30360 | 19953..20465 | - | 513 | WP_029697418 | phospholipase D family protein | - |
| AH61_RS30365 | 20477..21478 | - | 1002 | WP_001076634 | ATPase, T2SS/T4P/T4SS family | virB11 |
| AH61_RS30370 | 21529..22668 | - | 1140 | WP_014014945 | type IV secretion system protein VirB10 | virB10 |
| AH61_RS30375 | 22668..23555 | - | 888 | WP_014014946 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| AH61_RS30380 | 23566..24261 | - | 696 | WP_014014947 | type IV secretion system protein | virB8 |
| AH61_RS30385 | 24480..25517 | - | 1038 | WP_014014948 | type IV secretion system protein | virB6 |
| AH61_RS30390 | 25534..25776 | - | 243 | WP_000858958 | EexN family lipoprotein | - |
| AH61_RS30395 | 25786..26487 | - | 702 | WP_014014949 | type IV secretion system protein | virB5 |
| AH61_RS30400 | 26503..29076 | - | 2574 | WP_014014950 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| AH61_RS30405 | 29077..29394 | - | 318 | WP_000462629 | VirB3 family type IV secretion system protein | virB3 |
| AH61_RS30410 | 29434..29775 | - | 342 | WP_001121032 | TrbC/VirB2 family protein | virB2 |
| AH61_RS30415 | 29977..30759 | - | 783 | WP_014014952 | lytic transglycosylase domain-containing protein | virB1 |
| AH61_RS30420 | 30859..31176 | + | 318 | WP_000960955 | H-NS family nucleoid-associated regulatory protein | - |
| AH61_RS30425 | 31180..31530 | + | 351 | WP_014014953 | hypothetical protein | - |
| AH61_RS30430 | 31527..31865 | + | 339 | WP_000040982 | hypothetical protein | - |
| AH61_RS30435 | 31869..32177 | + | 309 | WP_000733764 | hypothetical protein | - |
| AH61_RS30440 | 32462..32647 | + | 186 | WP_014014955 | ribbon-helix-helix domain-containing protein | - |
| AH61_RS30445 | 32644..33252 | - | 609 | WP_042939603 | recombinase family protein | - |
Host bacterium
| ID | 1686 | GenBank | NZ_JAPZ01000355 |
| Plasmid name | pTR3 | Incompatibility group | - |
| Plasmid size | 33313 bp | Coordinate of oriT [Strand] | 4180..4262 [-] |
| Host baterium | Klebsiella pneumoniae JT4 |
Cargo genes
| Drug resistance gene | blaNDM-1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |