Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101197 |
| Name | oriT_pSM112_Rh04 |
| Organism | Rhizobium ruizarguesonis strain SM112 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SINL01000004 (21672..21712 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM112_Rh04
TCCAATGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
TCCAATGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file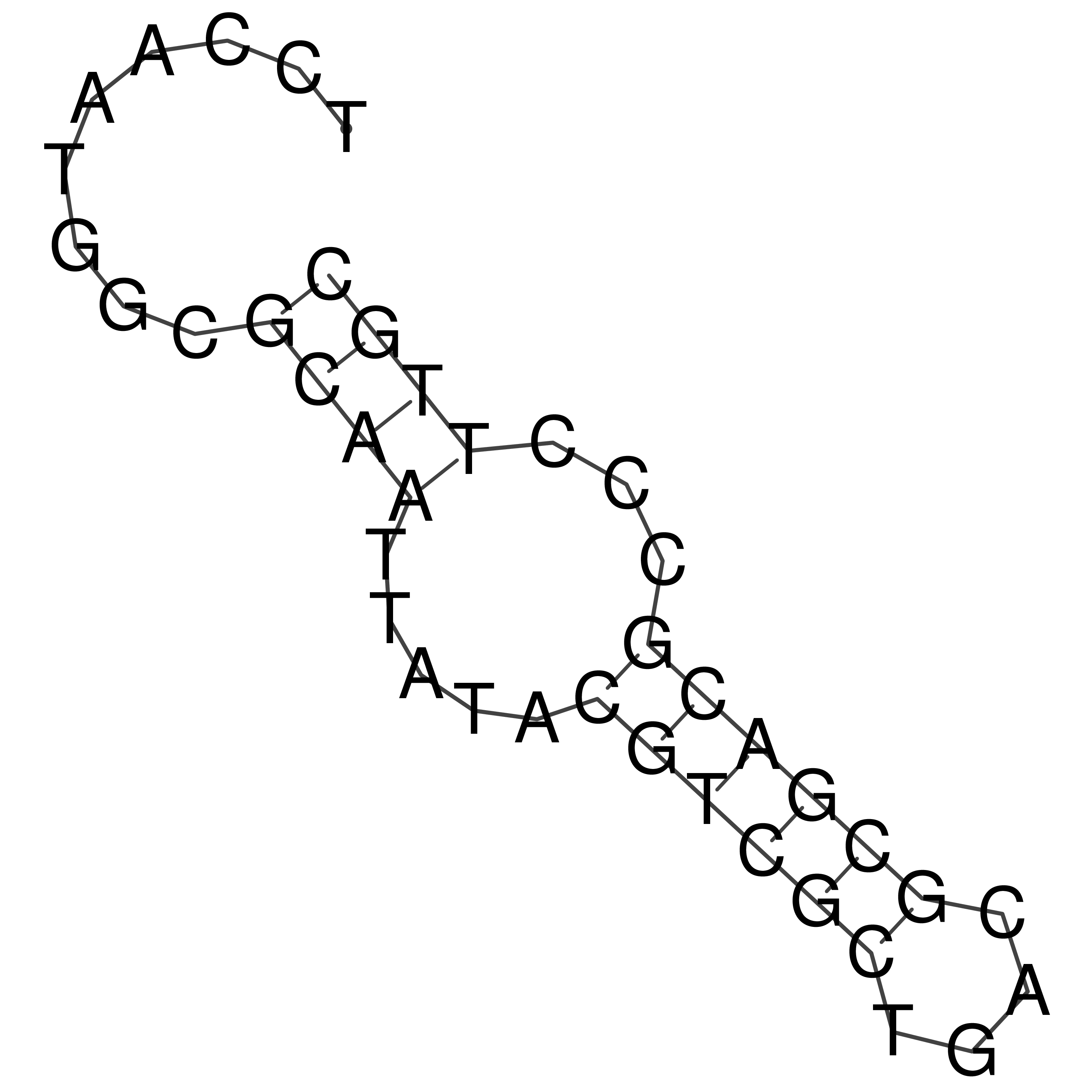
Relaxase
| ID | 1193 | GenBank | WP_130674195 |
| Name | traA_ELH60_RS36575_pSM112_Rh04 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122861.94 Da Isoelectric Point: 10.2104
>WP_130674195.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLHEEFVIPTGAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDSQLAKDLTIALPIELSAEQNVALVRDFVERHITSKGMVADWVYHDVPGN
PHVHLMTTLRPLTEDGFGAKKIAVASPDGNPMRNDAGKIVYELWAGSADDFNAVRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARTSAWSPKLERVELQQDRRAENARRIQRRPD
IVLDLITREKSVFDDRDVGKILHRYIDDAALFQNLMMRILQSPQALRLERDRIDFATGVRTPAKYTTREM
IKLEAEMVNRAIWLSDRASHGVREAALEVTFARHARLSDEQRTAIERVAGGERIAAVMGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNRIDARTVFVLDEAGMVSSRQM
AHFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRAQWMRDASLDLARGNVGKA
VDAYRAHGQVRGLDLKAQAVENLIADWNHDYDPSKSSLILAHLRRDVRMLNDMARAKLVERGIVADGFAF
KSEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVVALGEGDHRRVVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLVPILARRNAKETTLDYE
KSSFYGQALRFAEARGLHLMNVARTLARDRLEWTVRQKQKLADLGARLAAIGVKLGRAANAKTSPNLNAK
ESKPMVSGISTFSKSIDQSVEDKLAADPGLKKQWEDVSARFHLVYAQPESAFKAANVEAMLKDQAIAKTT
LAKLAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEVERKHETEERAVRRQVS
VDIPALSASAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELDGFAKAVSERFGERTFLGIASKDPSGE
TFKSVTAGMNPAQKAEVEIAWNSMRSIQQLSAHEKTTEALKQAETLRQTRSQGFSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLHEEFVIPTGAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDSQLAKDLTIALPIELSAEQNVALVRDFVERHITSKGMVADWVYHDVPGN
PHVHLMTTLRPLTEDGFGAKKIAVASPDGNPMRNDAGKIVYELWAGSADDFNAVRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARTSAWSPKLERVELQQDRRAENARRIQRRPD
IVLDLITREKSVFDDRDVGKILHRYIDDAALFQNLMMRILQSPQALRLERDRIDFATGVRTPAKYTTREM
IKLEAEMVNRAIWLSDRASHGVREAALEVTFARHARLSDEQRTAIERVAGGERIAAVMGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELRWNQGRNRIDARTVFVLDEAGMVSSRQM
AHFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRAQWMRDASLDLARGNVGKA
VDAYRAHGQVRGLDLKAQAVENLIADWNHDYDPSKSSLILAHLRRDVRMLNDMARAKLVERGIVADGFAF
KSEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVVALGEGDHRRVVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLVPILARRNAKETTLDYE
KSSFYGQALRFAEARGLHLMNVARTLARDRLEWTVRQKQKLADLGARLAAIGVKLGRAANAKTSPNLNAK
ESKPMVSGISTFSKSIDQSVEDKLAADPGLKKQWEDVSARFHLVYAQPESAFKAANVEAMLKDQAIAKTT
LAKLAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEVERKHETEERAVRRQVS
VDIPALSASAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELDGFAKAVSERFGERTFLGIASKDPSGE
TFKSVTAGMNPAQKAEVEIAWNSMRSIQQLSAHEKTTEALKQAETLRQTRSQGFSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 802 | GenBank | WP_024323836 |
| Name | traG_ELH60_RS36560_pSM112_Rh04 |
UniProt ID | _ |
| Length | 655 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 655 a.a. Molecular weight: 71050.85 Da Isoelectric Point: 8.7825
>WP_024323836.1 Ti-type conjugative transfer system protein TraG [Rhizobium ruizarguesonis]
MTANKIALFSVPIALMILVTIGMTGTEQWFSEFGKTETGRLILGRTGIALPYIASAATGIIFLFASAGST
NIKLAGWGVVGGSSATVVIAALRETVRLAAIFDHFPAVRSALSYADPATMIGAGAALMSAGFALRVALVG
NAAFARSEPRRIRGRRALHGEAEWMKSTEAGKLFPDTGGIVIGERYRVDKDIAAKQSFRADSVETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVLLDPSNEVAPMVIGHRTNAGRDVHILD
PRTPDTGFNALDWIGLHGRTKEEDIAAVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDRTEE
DDQTLRQVRKNLSEPEPKLRARLQSIYDNSRSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSESTFSTNDISAGNADVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGALKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
ENRFHSPARGDESAMDPETPMKLRG
MTANKIALFSVPIALMILVTIGMTGTEQWFSEFGKTETGRLILGRTGIALPYIASAATGIIFLFASAGST
NIKLAGWGVVGGSSATVVIAALRETVRLAAIFDHFPAVRSALSYADPATMIGAGAALMSAGFALRVALVG
NAAFARSEPRRIRGRRALHGEAEWMKSTEAGKLFPDTGGIVIGERYRVDKDIAAKQSFRADSVETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVLLDPSNEVAPMVIGHRTNAGRDVHILD
PRTPDTGFNALDWIGLHGRTKEEDIAAVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDRTEE
DDQTLRQVRKNLSEPEPKLRARLQSIYDNSRSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSESTFSTNDISAGNADVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGALKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
ENRFHSPARGDESAMDPETPMKLRG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 803 | GenBank | WP_024323902 |
| Name | t4cp2_ELH60_RS36765_pSM112_Rh04 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 91549.04 Da Isoelectric Point: 6.2219
>WP_024323902.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVSLRAFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEVSRQINAILSRLG
SGWMIQVEAVRLATVDYASAGKSHFPDRVTQAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YIYSDEDSRKQSYADTVLFIFRNAIREVEQYLANIISIRRMLTRETVERGGARVAHYDELLQFIRFCVTG
ENHPVRVPDIPMYLDWIATAELRHGLAPKVENRFLGVVAIDGLPSESWPGILNSLDAMPLGYRWSSRFIF
LDAEEARVRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALSMVAETEDAIAQASSQLVAYGYYTPVVVL
FESDSERLNEKTEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNSVWS
GNPRAPCPFYSPDSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLACGGDHYEIGGDEDTGLSFCPLSELSSEGDRAWASEWIESLVGMQGITITPDHRNAIS
RQIGLMDGASGRSISDFVSGVQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQTFEIEQVMNMGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTRDFYERIGFNERQIEIVATAIPKREYYVASPEGRRLFDMALGPLTL
SFVGASSKDDLKHIRALHGEHGPQWPLHWLQKRGIKDAASLLKDA
MVSLRAFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEVSRQINAILSRLG
SGWMIQVEAVRLATVDYASAGKSHFPDRVTQAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YIYSDEDSRKQSYADTVLFIFRNAIREVEQYLANIISIRRMLTRETVERGGARVAHYDELLQFIRFCVTG
ENHPVRVPDIPMYLDWIATAELRHGLAPKVENRFLGVVAIDGLPSESWPGILNSLDAMPLGYRWSSRFIF
LDAEEARVRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALSMVAETEDAIAQASSQLVAYGYYTPVVVL
FESDSERLNEKTEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNSVWS
GNPRAPCPFYSPDSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLACGGDHYEIGGDEDTGLSFCPLSELSSEGDRAWASEWIESLVGMQGITITPDHRNAIS
RQIGLMDGASGRSISDFVSGVQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQTFEIEQVMNMGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTRDFYERIGFNERQIEIVATAIPKREYYVASPEGRRLFDMALGPLTL
SFVGASSKDDLKHIRALHGEHGPQWPLHWLQKRGIKDAASLLKDA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 51452..60921
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH60_RS36705 (ELH60_36700) | 46916..47122 | + | 207 | WP_080642907 | hypothetical protein | - |
| ELH60_RS36710 (ELH60_36705) | 47242..47394 | - | 153 | Protein_42 | aminoglycoside phosphotransferase | - |
| ELH60_RS36715 (ELH60_36710) | 47446..49407 | - | 1962 | WP_130674184 | elongation factor G | - |
| ELH60_RS36720 (ELH60_36715) | 50247..50450 | - | 204 | Protein_44 | IS6 family transposase | - |
| ELH60_RS36725 (ELH60_36720) | 50716..51426 | + | 711 | WP_024323909 | autoinducer binding domain-containing protein | - |
| ELH60_RS36730 (ELH60_36725) | 51452..52732 | - | 1281 | WP_130674183 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH60_RS36735 (ELH60_36730) | 52747..53175 | - | 429 | WP_018068569 | conjugal transfer protein TrbH | - |
| ELH60_RS36740 (ELH60_36735) | 53178..53999 | - | 822 | WP_018068568 | P-type conjugative transfer protein TrbG | virB9 |
| ELH60_RS36745 (ELH60_36740) | 54016..54678 | - | 663 | WP_024323906 | conjugal transfer protein TrbF | virB8 |
| ELH60_RS36750 (ELH60_36745) | 54699..55847 | - | 1149 | WP_050574296 | P-type conjugative transfer protein TrbL | virB6 |
| ELH60_RS36755 (ELH60_36750) | 55874..56071 | - | 198 | WP_130674182 | entry exclusion protein TrbK | - |
| ELH60_RS36760 (ELH60_36755) | 56068..56868 | - | 801 | WP_130674181 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH60_RS36765 (ELH60_36760) | 56840..59287 | - | 2448 | WP_024323902 | conjugal transfer protein TrbE | virb4 |
| ELH60_RS36770 (ELH60_36765) | 59297..59596 | - | 300 | WP_024323901 | conjugal transfer protein TrbD | virB3 |
| ELH60_RS36775 (ELH60_36770) | 59586..59966 | - | 381 | WP_018068562 | TrbC/VirB2 family protein | virB2 |
| ELH60_RS36780 (ELH60_36775) | 59956..60921 | - | 966 | WP_024323900 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH60_RS36785 (ELH60_36780) | 60935..61561 | - | 627 | WP_130674180 | acyl-homoserine-lactone synthase TraI | - |
| ELH60_RS36790 (ELH60_36785) | 61926..63143 | + | 1218 | WP_075225817 | plasmid partitioning protein RepA | - |
| ELH60_RS36795 (ELH60_36790) | 63128..64168 | + | 1041 | WP_027689819 | plasmid partitioning protein RepB | - |
| ELH60_RS36800 (ELH60_36795) | 64407..65672 | + | 1266 | WP_130674179 | plasmid replication protein RepC | - |
Host bacterium
| ID | 1641 | GenBank | NZ_SINL01000004 |
| Plasmid name | pSM112_Rh04 | Incompatibility group | - |
| Plasmid size | 137764 bp | Coordinate of oriT [Strand] | 21672..21712 [+] |
| Host baterium | Rhizobium ruizarguesonis strain SM112 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nodA, nodI, nifT, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE |
| Anti-CRISPR | - |