Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101188 |
| Name | oriT_pSM153C_Rh08_Rh04 |
| Organism | Rhizobium ruizarguesonis strain SM153C |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIPY01000005 (91075..91115 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM153C_Rh08_Rh04
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file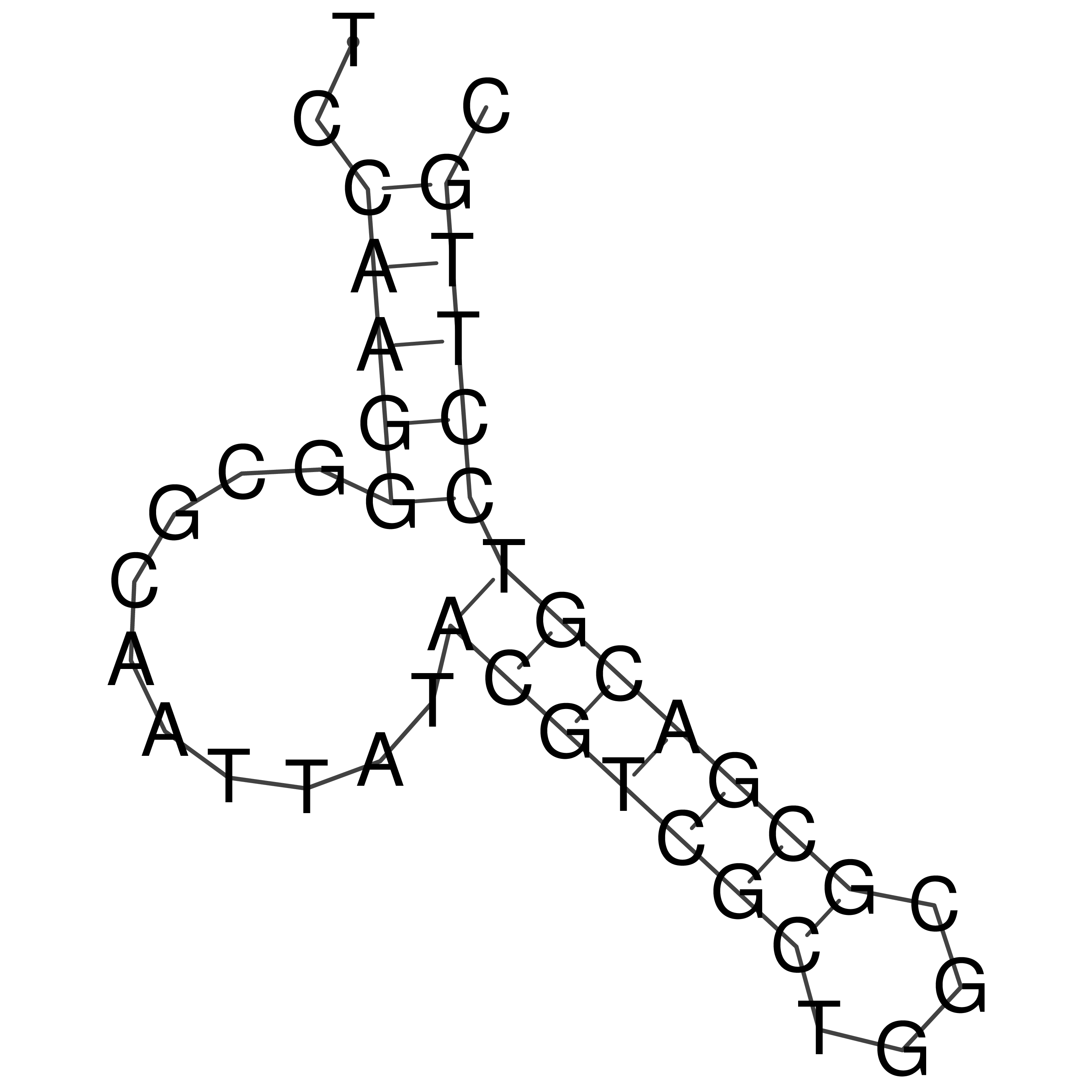
Relaxase
| ID | 1185 | GenBank | WP_130714113 |
| Name | traA_ELI25_RS32025_pSM153C_Rh08_Rh04 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123097.40 Da Isoelectric Point: 9.7427
>WP_130714113.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTVEQNIALMRDFVAGHITAQGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRRQVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KSALYRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKAIGLVRGSNKQTAPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAGDPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVKDQSVAQS
TLARIAGEPESFGALKGKAGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTVEQNIALMRDFVAGHITAQGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRRQVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KSALYRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKAIGLVRGSNKQTAPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAGDPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVKDQSVAQS
TLARIAGEPESFGALKGKAGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 788 | GenBank | WP_130714111 |
| Name | traG_ELI25_RS32010_pSM153C_Rh08_Rh04 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69549.58 Da Isoelectric Point: 9.5666
>WP_130714111.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVIVPTALMIPVVIGMTGIEQWLSGFGKTESARLAWGRTGIALPYVASAAIGVLWLFSSAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSGFMKTVPADKTVLAYLDPATSIGASTALLGACFALRVALIG
KAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSEHRGGAGRDVFILD
PRKPDTGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
NGQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGRKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMIGDAPKPA
MTINRIALVIVPTALMIPVVIGMTGIEQWLSGFGKTESARLAWGRTGIALPYVASAAIGVLWLFSSAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSGFMKTVPADKTVLAYLDPATSIGASTALLGACFALRVALIG
KAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSEHRGGAGRDVFILD
PRKPDTGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
NGQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGRKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMIGDAPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 789 | GenBank | WP_130747474 |
| Name | t4cp2_ELI25_RS32105_pSM153C_Rh08_Rh04 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91495.22 Da Isoelectric Point: 6.0219
>WP_130747474.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLLAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIQTVNYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YTYSDEVSRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELMQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDRVALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLKLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLCHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAASLLK
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLLAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIQTVNYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YTYSDEVSRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELMQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDRVALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLKLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLCHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAASLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 96243..110007
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI25_RS32030 (ELI25_32010) | 94510..95073 | + | 564 | WP_130714114 | conjugative transfer signal peptidase TraF | - |
| ELI25_RS32035 (ELI25_32015) | 95063..96226 | + | 1164 | WP_130714115 | conjugal transfer protein TraB | - |
| ELI25_RS32040 (ELI25_32020) | 96243..96860 | + | 618 | WP_130714116 | TraH family protein | virB1 |
| ELI25_RS32045 (ELI25_32025) | 96967..97832 | - | 866 | Protein_91 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELI25_RS32050 (ELI25_32030) | 97825..98442 | - | 618 | WP_130714117 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELI25_RS32055 (ELI25_32035) | 98684..98932 | - | 249 | WP_130714118 | helix-turn-helix transcriptional regulator | - |
| ELI25_RS32060 (ELI25_32040) | 99192..99482 | + | 291 | WP_130714119 | transcriptional repressor TraM | - |
| ELI25_RS32065 (ELI25_32045) | 99486..100190 | - | 705 | WP_130714120 | autoinducer binding domain-containing protein | - |
| ELI25_RS32070 (ELI25_32050) | 100502..101800 | - | 1299 | WP_130747468 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI25_RS32075 (ELI25_32055) | 101812..102252 | - | 441 | WP_130747469 | conjugal transfer protein TrbH | - |
| ELI25_RS32080 (ELI25_32060) | 102256..103068 | - | 813 | WP_130747470 | P-type conjugative transfer protein TrbG | virB9 |
| ELI25_RS32085 (ELI25_32065) | 103086..103748 | - | 663 | WP_130747471 | conjugal transfer protein TrbF | virB8 |
| ELI25_RS32090 (ELI25_32070) | 103769..104938 | - | 1170 | WP_130747472 | P-type conjugative transfer protein TrbL | virB6 |
| ELI25_RS32095 (ELI25_32075) | 104932..105132 | - | 201 | WP_130747477 | entry exclusion protein TrbK | - |
| ELI25_RS32100 (ELI25_32080) | 105129..105926 | - | 798 | WP_130747473 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI25_RS32105 (ELI25_32085) | 105904..108354 | - | 2451 | WP_130747474 | conjugal transfer protein TrbE | virb4 |
| ELI25_RS32110 (ELI25_32090) | 108365..108664 | - | 300 | WP_130714129 | conjugal transfer protein TrbD | virB3 |
| ELI25_RS32115 (ELI25_32095) | 108657..109040 | - | 384 | WP_130714130 | TrbC/VirB2 family protein | virB2 |
| ELI25_RS32120 (ELI25_32100) | 109030..110007 | - | 978 | WP_130746550 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI25_RS32125 (ELI25_32105) | 110004..110642 | - | 639 | WP_130714132 | acyl-homoserine-lactone synthase | - |
| ELI25_RS32130 (ELI25_32110) | 111019..112242 | + | 1224 | WP_165411169 | plasmid partitioning protein RepA | - |
| ELI25_RS32135 (ELI25_32115) | 112239..113276 | + | 1038 | WP_130714134 | plasmid partitioning protein RepB | - |
| ELI25_RS32140 (ELI25_32120) | 113466..114680 | + | 1215 | WP_130714135 | plasmid replication protein RepC | - |
Host bacterium
| ID | 1632 | GenBank | NZ_SIPY01000005 |
| Plasmid name | pSM153C_Rh08_Rh04 | Incompatibility group | - |
| Plasmid size | 131997 bp | Coordinate of oriT [Strand] | 91075..91115 [+] |
| Host baterium | Rhizobium ruizarguesonis strain SM153C |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD |
| Anti-CRISPR | - |