Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101178 |
| Name | oriT_pSM141A_Rh17 |
| Organism | Rhizobium ruizarguesonis strain SM141A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOX01000008 (29964..30013 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pSM141A_Rh17
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file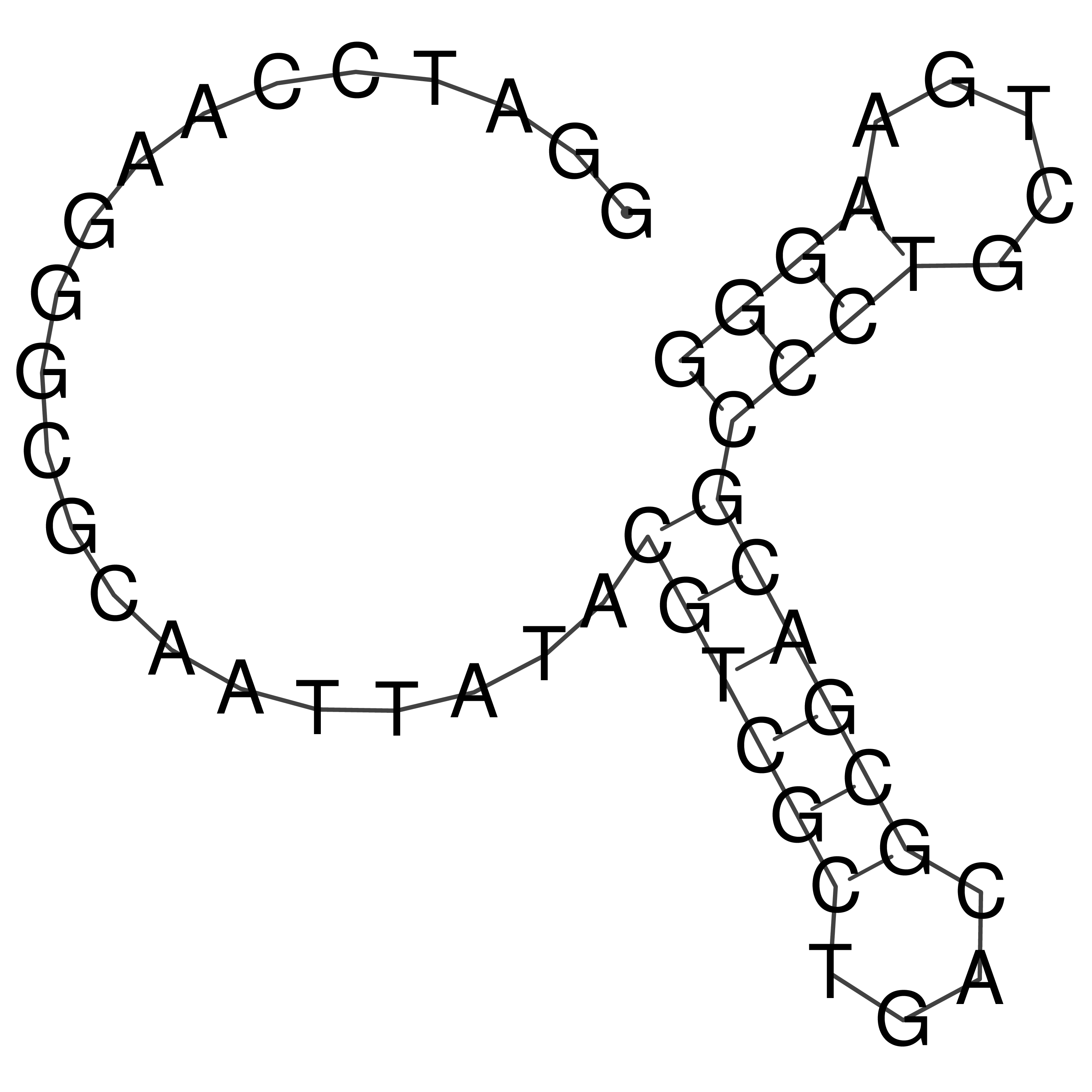
Relaxase
| ID | 1175 | GenBank | WP_115039354 |
| Name | traA_ELH98_RS29865_pSM141A_Rh17 |
UniProt ID | _ |
| Length | 1197 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1197 a.a. Molecular weight: 131867.79 Da Isoelectric Point: 9.4235
>WP_115039354.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPDGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGLAEEKVALERLELQEERRAENARRIQRNPEI
VVDLITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELI
RLEAQMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVIVLDEAGMVSSRQMA
RFVEAVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDLVRTGWTRDEAITALIAEWDHKYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMA
RLHERLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTTSVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKAVNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAK
RAAAARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPDGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKAASGNGLAEEKVALERLELQEERRAENARRIQRNPEI
VVDLITREKSVFDERDIAKVLYRYIDDAALFQNLMARLLQSPQTLRLDRERMDLVTGVRAPAKYTTRELI
RLEAQMANQAIWLSQRSSHRVNRTVLSGIVSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
AREAWEAAGYRVVGGALAGKAADGLEKEAGIASRTLSAWELRWDQERDRLDEKSVIVLDEAGMVSSRQMA
RFVEAVTLSGAKLVLVGDPEQLQPIEAGAAFRAIAERIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDLVRTGWTRDEAITALIAEWDHKYDPAKSTLILAHRRVDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQFAAGDQIVFLKNEGSLGVKNGMLALVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFANGQERTVWGVDLARAMDASDSRIGDRIGLKHVGSQRVTLPNGTEVDRNSWKVVPIQELAMA
RLHERLSRAGSKETTLDYQDASHYRAALRFAEARGLHLMNVARTIAHDQLQWTVRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTTSVIKEAKPMVFGTTTFPRSIGQAVEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKAVNVDAMLANGTVAATTIVRIAEQPESFGALKGKTGLFAGSAEKQARDTALVNAPALARDLQGFIAK
RAAAARRYEDEERAVRAQLSLDIPALSASGKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 769 | GenBank | WP_115039372 |
| Name | t4cp2_ELH98_RS29780_pSM141A_Rh17 |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 90978.39 Da Isoelectric Point: 5.8031
>WP_115039372.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCGVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPDHWLIERGINRNE
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCGVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPDHWLIERGINRNE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 770 | GenBank | WP_130749266 |
| Name | traG_ELH98_RS29880_pSM141A_Rh17 |
UniProt ID | _ |
| Length | 650 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 650 a.a. Molecular weight: 70585.41 Da Isoelectric Point: 9.7031
>WP_130749266.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTPKRILVAGIPAVAMLAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
MTPKRILVAGIPAVAMLAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SAPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9338..24567
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH98_RS29750 (ELH98_29745) | 5292..6536 | - | 1245 | WP_115039376 | plasmid replication protein RepC | - |
| ELH98_RS29755 (ELH98_29750) | 6694..7686 | - | 993 | WP_064251333 | plasmid partitioning protein RepB | - |
| ELH98_RS29760 (ELH98_29755) | 7768..8958 | - | 1191 | WP_018009855 | plasmid partitioning protein RepA | - |
| ELH98_RS29765 (ELH98_29760) | 9338..10303 | + | 966 | WP_028744194 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH98_RS29770 (ELH98_29765) | 10293..10685 | + | 393 | WP_115039374 | TrbC/VirB2 family protein | virB2 |
| ELH98_RS29775 (ELH98_29770) | 10678..10977 | + | 300 | WP_011654761 | conjugal transfer protein TrbD | virB3 |
| ELH98_RS29780 (ELH98_29775) | 10988..13423 | + | 2436 | WP_115039372 | conjugal transfer protein TrbE | virb4 |
| ELH98_RS29785 (ELH98_29780) | 13416..14225 | + | 810 | WP_011654759 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH98_RS29790 (ELH98_29785) | 14222..14422 | + | 201 | WP_028744197 | entry exclusion protein TrbK | - |
| ELH98_RS29795 (ELH98_29790) | 14416..15597 | + | 1182 | WP_028744198 | P-type conjugative transfer protein TrbL | virB6 |
| ELH98_RS29800 (ELH98_29795) | 15619..16281 | + | 663 | WP_115039370 | conjugal transfer protein TrbF | virB8 |
| ELH98_RS29805 (ELH98_29800) | 16333..17127 | + | 795 | Protein_15 | P-type conjugative transfer protein TrbG | - |
| ELH98_RS29810 (ELH98_29805) | 17131..17577 | + | 447 | WP_115039367 | conjugal transfer protein TrbH | - |
| ELH98_RS29815 (ELH98_29810) | 17593..18894 | + | 1302 | WP_115039365 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH98_RS29820 (ELH98_29815) | 19095..19394 | + | 300 | WP_027690597 | transcriptional repressor TraM | - |
| ELH98_RS29825 (ELH98_29820) | 19395..20099 | - | 705 | WP_115039418 | autoinducer binding domain-containing protein | - |
| ELH98_RS29830 (ELH98_29825) | 20382..21998 | + | 1617 | WP_115039363 | hypothetical protein | - |
| ELH98_RS29835 (ELH98_29830) | 21988..22983 | + | 996 | WP_115039362 | dienelactone hydrolase-related enzyme | - |
| ELH98_RS29840 (ELH98_29835) | 23133..23312 | + | 180 | WP_018010295 | hypothetical protein | - |
| ELH98_RS29845 (ELH98_29840) | 23369..23926 | + | 558 | WP_245486992 | hypothetical protein | - |
| ELH98_RS29850 (ELH98_29845) | 23950..24567 | - | 618 | WP_115039357 | TraH family protein | virB1 |
| ELH98_RS29855 (ELH98_29850) | 24583..25749 | - | 1167 | WP_115039355 | conjugal transfer protein TraB | - |
| ELH98_RS29860 (ELH98_29855) | 25739..26302 | - | 564 | WP_050595854 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 1622 | GenBank | NZ_SIOX01000008 |
| Plasmid name | pSM141A_Rh17 | Incompatibility group | - |
| Plasmid size | 252224 bp | Coordinate of oriT [Strand] | 29964..30013 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM141A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |