Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101173 |
| Name | oriT_pSM165B_Rh15 |
| Organism | Rhizobium ruizarguesonis strain SM165B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQT01000008 (787..828 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 22..27, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
>oriT_pSM165B_Rh15
GGCTCCAAGGGCGCAATTATACGTCGCGATGCGACGCGTTGC
GGCTCCAAGGGCGCAATTATACGTCGCGATGCGACGCGTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file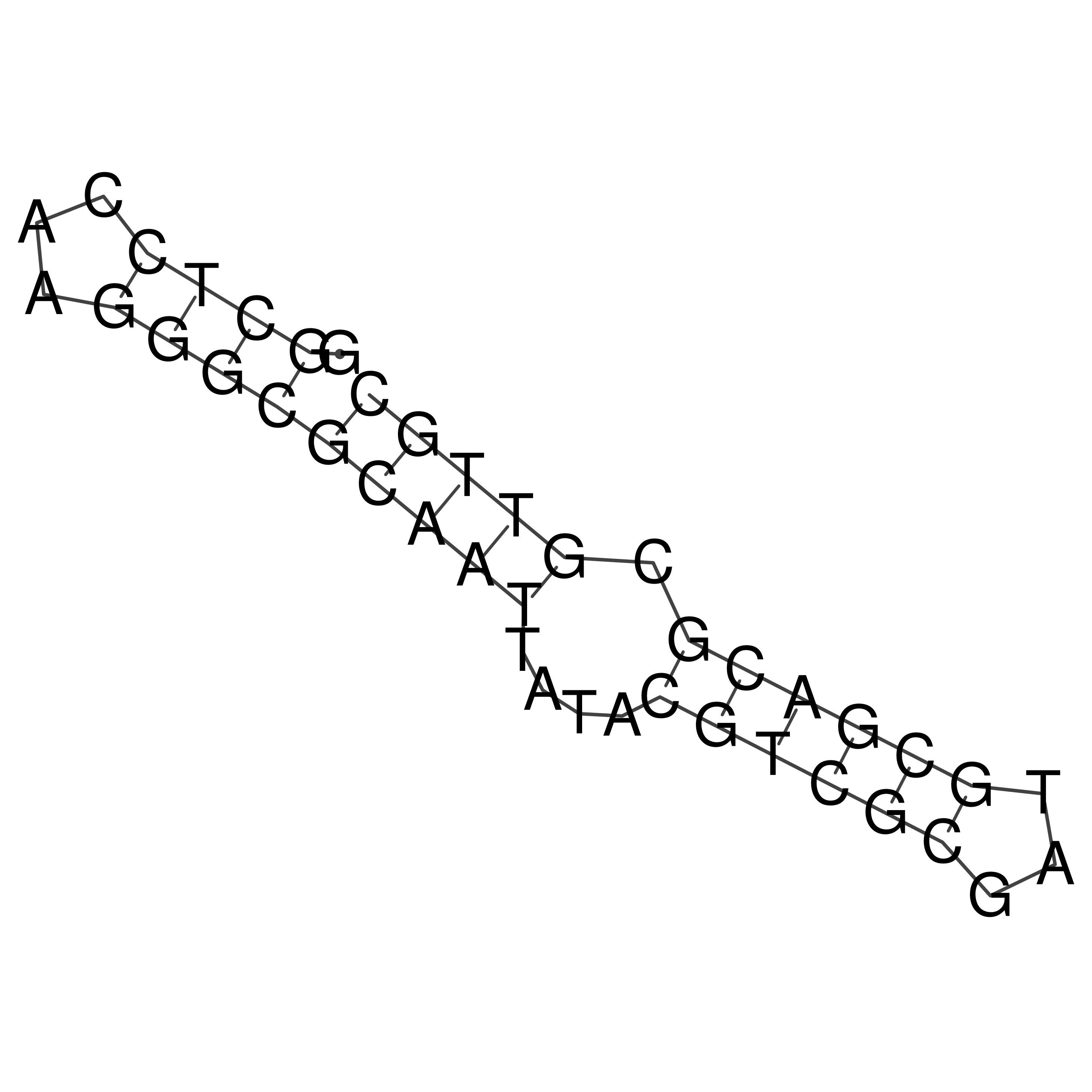
Relaxase
| ID | 1170 | GenBank | WP_130738477 |
| Name | traA_ELI46_RS30015_pSM165B_Rh15 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123330.67 Da Isoelectric Point: 9.8625
>WP_130738477.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPSDAPEWVRSLIADR
SVSGAVEAFWNRVEAFEKRADAQLARDLTIALPLELSAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHVHLMTTLRPLSDDGFGSKKIAVIGEDGQPVRTKSGKIFYELWAGSTDDFNVLRNGWFERLNHHLALGG
VDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAEQRGVRPELERIELNEQRRSENTRRILRNPAIVLDL
ITREKSVFDERDIAKVLHRYIDDPALFQQLMLKIILNPDVLRLQRETIDFTTGDKLLARFSTRAMIRLEA
TMARQAMWLSNREGRGVSHSALDATFRRHERLSDEQKAAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDGLDDKTIFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRVGYAELETIYRQREDWMRKASLDLARGNVEKALSAYG
ANVRITGERLKAEAVERLIADWNHDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGMVGEGHLFRTADG
QRRFDAGDQIVFLKNEASLGVKNGMIGHVVESAANGIVATVGEGDQRRQVFIEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLQLYYGTRSFYFNGGLAKVLSRKNAKETTLDYASGQLY
REALRFSENRGLHIVQVARTLVRDRLDWTLRQKAKLVDLSERLAAFAARLGFTQSPTTQTKKEAPAMVAG
IKTFSGSVADTVGGRLDADPSLKRQWEEVSARFAYVFADPETAFRGMNFDAVLADKEVAKQILKKLEAEP
ETIGPLKGKTGILAAKSEREARRVAEVNVPALTRDLEQYLHIRETAATRLQTDEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNKAVTERFGERTLLSNAAREPSGKLYEKLSEG
MKPEQKEQLKQAWPVMRTAQQLAAHERTVHSLKLAEEQRLAQRQTPVLKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPSDAPEWVRSLIADR
SVSGAVEAFWNRVEAFEKRADAQLARDLTIALPLELSAEQNIALVRDFVEKHILAKGMVADWVYHDNPGN
PHVHLMTTLRPLSDDGFGSKKIAVIGEDGQPVRTKSGKIFYELWAGSTDDFNVLRNGWFERLNHHLALGG
VDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAEQRGVRPELERIELNEQRRSENTRRILRNPAIVLDL
ITREKSVFDERDIAKVLHRYIDDPALFQQLMLKIILNPDVLRLQRETIDFTTGDKLLARFSTRAMIRLEA
TMARQAMWLSNREGRGVSHSALDATFRRHERLSDEQKAAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDGLDDKTIFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRVGYAELETIYRQREDWMRKASLDLARGNVEKALSAYG
ANVRITGERLKAEAVERLIADWNHDYDQTKTTLILAHLRRDVRMLNVMAREKLVERGMVGEGHLFRTADG
QRRFDAGDQIVFLKNEASLGVKNGMIGHVVESAANGIVATVGEGDQRRQVFIEQRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLQLYYGTRSFYFNGGLAKVLSRKNAKETTLDYASGQLY
REALRFSENRGLHIVQVARTLVRDRLDWTLRQKAKLVDLSERLAAFAARLGFTQSPTTQTKKEAPAMVAG
IKTFSGSVADTVGGRLDADPSLKRQWEEVSARFAYVFADPETAFRGMNFDAVLADKEVAKQILKKLEAEP
ETIGPLKGKTGILAAKSEREARRVAEVNVPALTRDLEQYLHIRETAATRLQTDEQALRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNKAVTERFGERTLLSNAAREPSGKLYEKLSEG
MKPEQKEQLKQAWPVMRTAQQLAAHERTVHSLKLAEEQRLAQRQTPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 761 | GenBank | WP_130738501 |
| Name | t4cp2_ELI46_RS30255_pSM165B_Rh15 |
UniProt ID | _ |
| Length | 820 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 820 a.a. Molecular weight: 91490.54 Da Isoelectric Point: 6.2201
>WP_130738501.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRVPTGDYPTEEASHFPDPVTRAIDAERRAHFQKERGHYESRHALILTWRPPEPRRSGLTR
YVYSDVGSRSATYADKARTSFLTSIREVEQYLANVVSIRRMMTRETPERGGHRVARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIIL
FDEDQIRLQEKCEAVRRLIQAEGFGARIETINATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSIWC
GSPIAPCPFYPVGAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYAGAQIFA
FDKGGSMLPLTLGLGGDHYQIGGDAQEAGEGRALSFCPLAELSTDGDRAFASEWIEMLVALQGVTITPDY
RNAISRQVGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEADGLALGAFQCFEIEELMN
MGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDA
ERSGIIDVLKESCPAKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKRDYYVVSPEGRRLFDMAL
GPVALSFVGASGKEDLKRIRALHLEHGASWPLHWLQQRGIAHADTLIPIH
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRVPTGDYPTEEASHFPDPVTRAIDAERRAHFQKERGHYESRHALILTWRPPEPRRSGLTR
YVYSDVGSRSATYADKARTSFLTSIREVEQYLANVVSIRRMMTRETPERGGHRVARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIIL
FDEDQIRLQEKCEAVRRLIQAEGFGARIETINATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSIWC
GSPIAPCPFYPVGAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYAGAQIFA
FDKGGSMLPLTLGLGGDHYQIGGDAQEAGEGRALSFCPLAELSTDGDRAFASEWIEMLVALQGVTITPDY
RNAISRQVGLMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEADGLALGAFQCFEIEELMN
MGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDA
ERSGIIDVLKESCPAKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKRDYYVVSPEGRRLFDMAL
GPVALSFVGASGKEDLKRIRALHLEHGASWPLHWLQQRGIAHADTLIPIH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 47640..57313
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI46_RS30205 (ELI46_30205) | 44230..45423 | - | 1194 | WP_130738496 | efflux RND transporter periplasmic adaptor subunit | - |
| ELI46_RS30210 (ELI46_30210) | 45645..47009 | - | 1365 | WP_130692398 | HAMP domain-containing sensor histidine kinase | - |
| ELI46_RS30215 (ELI46_30215) | 46996..47209 | - | 214 | Protein_43 | winged helix-turn-helix domain-containing protein | - |
| ELI46_RS41900 | 47397..47522 | - | 126 | WP_281032928 | hypothetical protein | - |
| ELI46_RS30220 (ELI46_30220) | 47640..48947 | - | 1308 | WP_130738497 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI46_RS30225 (ELI46_30225) | 48960..49439 | - | 480 | WP_115039455 | conjugal transfer protein TrbH | - |
| ELI46_RS30230 (ELI46_30230) | 49439..50260 | - | 822 | WP_197726622 | P-type conjugative transfer protein TrbG | virB9 |
| ELI46_RS30235 (ELI46_30235) | 50311..50973 | - | 663 | WP_112497059 | conjugal transfer protein TrbF | virB8 |
| ELI46_RS30240 (ELI46_30240) | 50988..52178 | - | 1191 | WP_130738498 | P-type conjugative transfer protein TrbL | virB6 |
| ELI46_RS30245 (ELI46_30245) | 52172..52399 | - | 228 | WP_130738499 | entry exclusion protein TrbK | - |
| ELI46_RS30250 (ELI46_30250) | 52396..53205 | - | 810 | WP_130738500 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI46_RS30255 (ELI46_30255) | 53177..55639 | - | 2463 | WP_130738501 | conjugal transfer protein TrbE | virb4 |
| ELI46_RS30260 (ELI46_30260) | 55650..55949 | - | 300 | WP_130738502 | conjugal transfer protein TrbD | virB3 |
| ELI46_RS30265 (ELI46_30265) | 55942..56352 | - | 411 | WP_130738503 | conjugal transfer pilin TrbC | virB2 |
| ELI46_RS30270 (ELI46_30270) | 56342..57313 | - | 972 | WP_130738504 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI46_RS30275 (ELI46_30275) | 57310..57945 | - | 636 | WP_130738505 | acyl-homoserine-lactone synthase | - |
| ELI46_RS30280 (ELI46_30280) | 58322..59512 | + | 1191 | WP_130738506 | plasmid partitioning protein RepA | - |
| ELI46_RS30285 (ELI46_30285) | 59597..60592 | + | 996 | WP_130738507 | plasmid partitioning protein RepB | - |
| ELI46_RS30290 (ELI46_30290) | 60749..61993 | + | 1245 | WP_130738508 | plasmid replication protein RepC | - |
Host bacterium
| ID | 1617 | GenBank | NZ_SIQT01000008 |
| Plasmid name | pSM165B_Rh15 | Incompatibility group | - |
| Plasmid size | 86303 bp | Coordinate of oriT [Strand] | 787..828 [+] |
| Host baterium | Rhizobium ruizarguesonis strain SM165B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |