Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101172 |
| Name | oriT_pSM165B_Rh07 |
| Organism | Rhizobium ruizarguesonis strain SM165B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQT01000006 (29602..29635 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM165B_Rh07
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file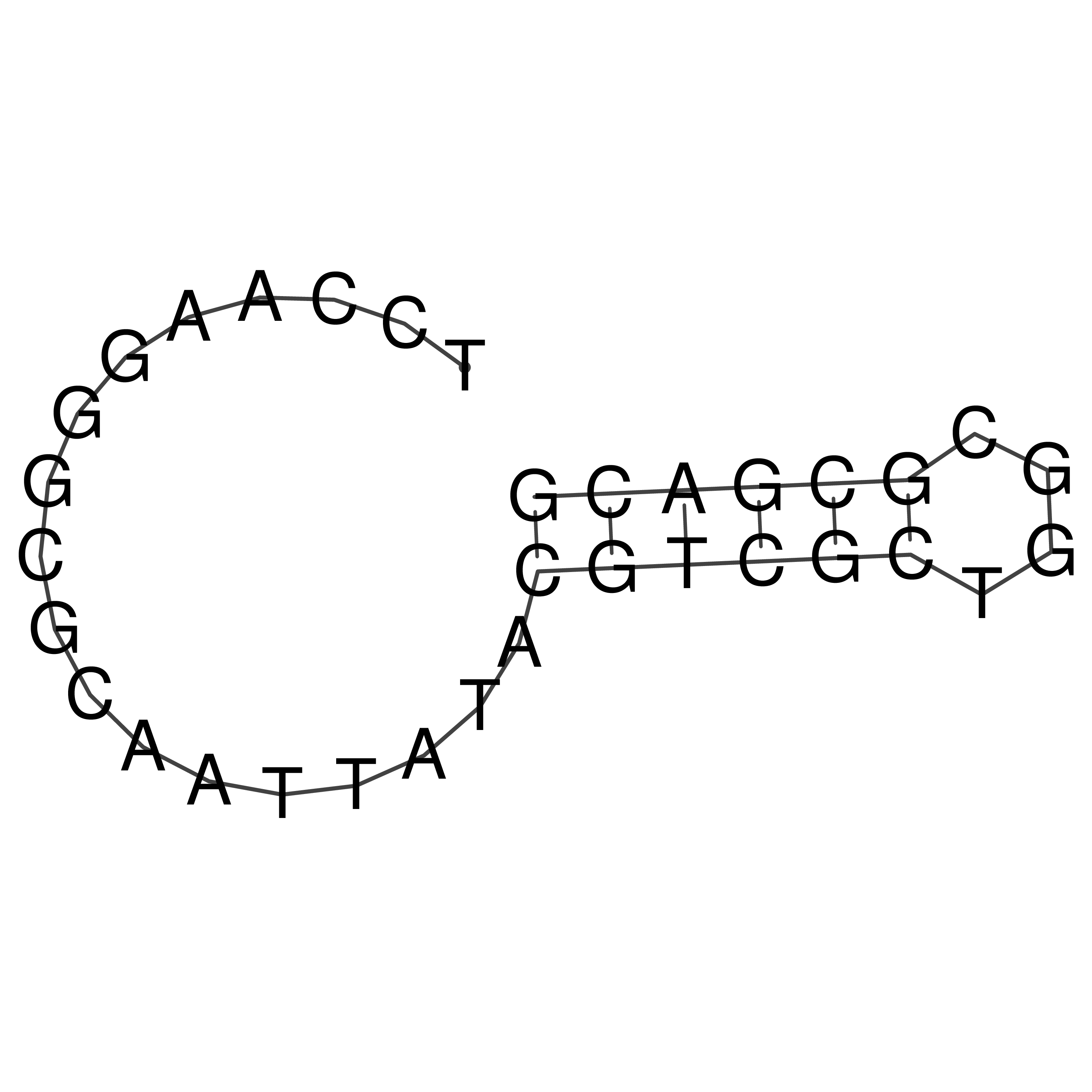
Relaxase
| ID | 1169 | GenBank | WP_130738440 |
| Name | traA_ELI46_RS29635_pSM165B_Rh07 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122984.17 Da Isoelectric Point: 9.7988
>WP_130738440.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAVPHFSVSVVARGSGHSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNISLMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGGTEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRALWLSGRASHGVRDAVLQATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDRCVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVFVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRQVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIARDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKVINVDTMLKDQSVAQA
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTGG
KTFETVTAGMTAGQKAEVQSAWNAMRTVQQLGSHERTTEAHKQAETLQTRSQGLSLK
MAVPHFSVSVVARGSGHSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNISLMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGGTEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRALWLSGRASHGVRDAVLQATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDRCVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVFVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLAKVLEAAPGRIVAEIGEGEHRRQVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETSLDYE
KSALYRQALRFAEARGLHLMNVARTIARDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIPDTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKVINVDTMLKDQSVAQA
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTGG
KTFETVTAGMTAGQKAEVQSAWNAMRTVQQLGSHERTTEAHKQAETLQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 759 | GenBank | WP_130738437 |
| Name | t4cp2_ELI46_RS29550_pSM165B_Rh07 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91757.43 Da Isoelectric Point: 5.8099
>WP_130738437.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKYV
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKYV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 760 | GenBank | WP_130738441 |
| Name | traG_ELI46_RS29650_pSM165B_Rh07 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69609.91 Da Isoelectric Point: 8.9733
>WP_130738441.1 Ti-type conjugative transfer system protein TraG [Rhizobium ruizarguesonis]
MTVNRIALAVVPTVLMILVVVGMTGIEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGVLLLFATAGAI
NIKQAGWGVLAGCCGTILIAVTRETMRLATFTKTLHADKSFLAYLDPATAIGASVALMCACFALRVVLVG
NAAFARPEPKRINGKRALHGEADWMKLTEAAKLFPDRGGIVIGERYRVDKDSVGARAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDVGFNVLDWVGCFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVSLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNIDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETAEYISKRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPEPA
MTVNRIALAVVPTVLMILVVVGMTGIEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGVLLLFATAGAI
NIKQAGWGVLAGCCGTILIAVTRETMRLATFTKTLHADKSFLAYLDPATAIGASVALMCACFALRVVLVG
NAAFARPEPKRINGKRALHGEADWMKLTEAAKLFPDRGGIVIGERYRVDKDSVGARAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDVGFNVLDWVGCFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVSLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNIDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETAEYISKRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPEPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 10288..24464
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI46_RS29510 (ELI46_29510) | 5819..6841 | - | 1023 | WP_128408794 | plasmid partitioning protein RepB | - |
| ELI46_RS29515 (ELI46_29515) | 6838..8052 | - | 1215 | WP_027691186 | plasmid partitioning protein RepA | - |
| ELI46_RS29520 (ELI46_29520) | 8669..8923 | - | 255 | WP_130692270 | IS66 family insertion sequence element accessory protein TnpB | - |
| ELI46_RS41450 | 8964..9176 | - | 213 | WP_245458276 | hypothetical protein | - |
| ELI46_RS29530 (ELI46_29530) | 9654..10277 | + | 624 | WP_130738434 | acyl-homoserine-lactone synthase | - |
| ELI46_RS29535 (ELI46_29535) | 10288..11265 | + | 978 | WP_130738435 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI46_RS29540 (ELI46_29540) | 11255..11638 | + | 384 | WP_130738436 | TrbC/VirB2 family protein | virB2 |
| ELI46_RS29545 (ELI46_29545) | 11631..11930 | + | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELI46_RS29550 (ELI46_29550) | 11941..14397 | + | 2457 | WP_130738437 | conjugal transfer protein TrbE | virb4 |
| ELI46_RS29555 (ELI46_29555) | 14369..15172 | + | 804 | WP_128408799 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI46_RS29560 (ELI46_29560) | 15169..15369 | + | 201 | WP_130680525 | entry exclusion protein TrbK | - |
| ELI46_RS29565 (ELI46_29565) | 15363..16544 | + | 1182 | WP_130738438 | P-type conjugative transfer protein TrbL | virB6 |
| ELI46_RS29570 (ELI46_29570) | 16565..17227 | + | 663 | WP_128408802 | conjugal transfer protein TrbF | virB8 |
| ELI46_RS29575 (ELI46_29575) | 17245..18057 | + | 813 | WP_130680527 | P-type conjugative transfer protein TrbG | virB9 |
| ELI46_RS29580 (ELI46_29580) | 18061..18501 | + | 441 | WP_130680528 | conjugal transfer protein TrbH | - |
| ELI46_RS29585 (ELI46_29585) | 18513..19808 | + | 1296 | WP_130680529 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI46_RS29590 (ELI46_29590) | 19993..20733 | + | 741 | WP_130680530 | LuxR family transcriptional regulator | - |
| ELI46_RS29595 (ELI46_29595) | 20840..21544 | + | 705 | WP_130680531 | autoinducer binding domain-containing protein | - |
| ELI46_RS29600 (ELI46_29600) | 21548..21838 | - | 291 | WP_130680532 | transcriptional repressor TraM | - |
| ELI46_RS29605 (ELI46_29605) | 22130..22378 | + | 249 | WP_130680533 | helix-turn-helix transcriptional regulator | - |
| ELI46_RS29610 (ELI46_29610) | 22499..23131 | + | 633 | WP_130680534 | DUF433 domain-containing protein | - |
| ELI46_RS29615 (ELI46_29615) | 23128..23502 | + | 375 | WP_130680535 | DUF5615 family PIN-like protein | - |
| ELI46_RS29620 (ELI46_29620) | 23844..24464 | - | 621 | WP_130738439 | TraH family protein | virB1 |
| ELI46_RS29625 (ELI46_29625) | 24481..25647 | - | 1167 | WP_130680551 | conjugal transfer protein TraB | - |
| ELI46_RS29630 (ELI46_29630) | 25637..26134 | - | 498 | WP_245484661 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 1616 | GenBank | NZ_SIQT01000006 |
| Plasmid name | pSM165B_Rh07 | Incompatibility group | - |
| Plasmid size | 44270 bp | Coordinate of oriT [Strand] | 29602..29635 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM165B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |