Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101168 |
| Name | oriT_pSM153A_Rh08 |
| Organism | Rhizobium ruizarguesonis strain SM153A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIPX01000007 (108459..108492 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM153A_Rh08
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file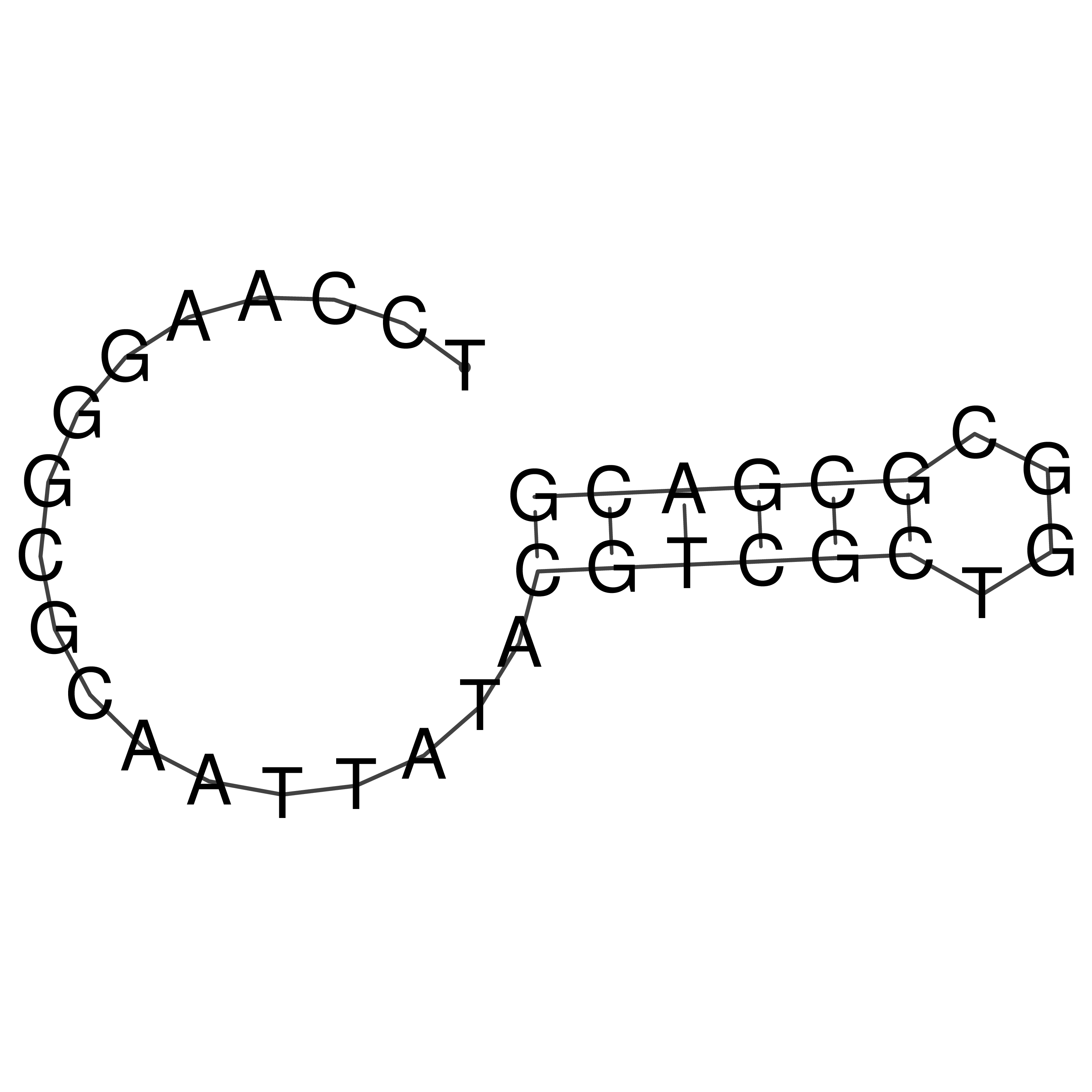
Relaxase
| ID | 1165 | GenBank | WP_130657460 |
| Name | traA_ELI24_RS32280_pSM153A_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122987.83 Da Isoelectric Point: 9.6219
>WP_130657460.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 751 | GenBank | WP_065276635 |
| Name | t4cp2_ELI24_RS32190_pSM153A_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91588.38 Da Isoelectric Point: 5.9977
>WP_065276635.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 752 | GenBank | WP_130657461 |
| Name | traG_ELI24_RS32295_pSM153A_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69518.76 Da Isoelectric Point: 8.9386
>WP_130657461.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 88188..103323
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI24_RS32155 (ELI24_32175) | 83515..84729 | - | 1215 | WP_065276629 | plasmid replication protein RepC | - |
| ELI24_RS32160 (ELI24_32180) | 84919..85956 | - | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELI24_RS32165 (ELI24_32185) | 85953..87176 | - | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELI24_RS32170 (ELI24_32190) | 87553..88191 | + | 639 | WP_130660092 | acyl-homoserine-lactone synthase | - |
| ELI24_RS32175 (ELI24_32195) | 88188..89165 | + | 978 | WP_065276632 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI24_RS32180 (ELI24_32200) | 89155..89538 | + | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELI24_RS32185 (ELI24_32205) | 89531..89830 | + | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELI24_RS32190 (ELI24_32210) | 89841..92291 | + | 2451 | WP_065276635 | conjugal transfer protein TrbE | virb4 |
| ELI24_RS32195 (ELI24_32215) | 92269..93066 | + | 798 | WP_130657453 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI24_RS32200 (ELI24_32220) | 93063..93263 | + | 201 | WP_130676499 | entry exclusion protein TrbK | - |
| ELI24_RS32205 (ELI24_32225) | 93257..94438 | + | 1182 | WP_065276638 | P-type conjugative transfer protein TrbL | virB6 |
| ELI24_RS32210 (ELI24_32230) | 94459..95121 | + | 663 | WP_130676500 | conjugal transfer protein TrbF | virB8 |
| ELI24_RS32215 (ELI24_32235) | 95139..95951 | + | 813 | WP_065276640 | P-type conjugative transfer protein TrbG | virB9 |
| ELI24_RS32220 (ELI24_32240) | 95955..96389 | + | 435 | WP_130657454 | conjugal transfer protein TrbH | - |
| ELI24_RS32225 (ELI24_32245) | 96401..97699 | + | 1299 | WP_130660093 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI24_RS32230 (ELI24_32250) | 98010..98714 | + | 705 | WP_130654288 | autoinducer binding domain-containing protein | - |
| ELI24_RS32235 (ELI24_32255) | 98718..99041 | - | 324 | WP_130654289 | transcriptional repressor TraM | - |
| ELI24_RS32240 (ELI24_32260) | 99264..99506 | + | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
| ELI24_RS40560 (ELI24_32265) | 99609..100040 | + | 432 | Protein_102 | hypothetical protein | - |
| ELI24_RS32250 (ELI24_32270) | 100131..100748 | + | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELI24_RS32255 (ELI24_32275) | 100741..101220 | + | 480 | Protein_104 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELI24_RS32260 (ELI24_32280) | 101217..102581 | + | 1365 | Protein_105 | IS66 family transposase | - |
| ELI24_RS32265 (ELI24_32285) | 102706..103323 | - | 618 | WP_130657457 | TraH family protein | virB1 |
| ELI24_RS32270 (ELI24_32290) | 103340..104503 | - | 1164 | WP_130657458 | conjugal transfer protein TraB | - |
| ELI24_RS32275 (ELI24_32295) | 104493..105056 | - | 564 | WP_130657459 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 1612 | GenBank | NZ_SIPX01000007 |
| Plasmid name | pSM153A_Rh08 | Incompatibility group | - |
| Plasmid size | 269002 bp | Coordinate of oriT [Strand] | 108459..108492 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM153A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, fixG, fixQ, fixO, fixN, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifT, nodJ, nodI, nodC, nodA, nodF, nodE, nodM |
| Anti-CRISPR | - |