Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101165 |
| Name | oriT_pSM107_Rh22 |
| Organism | Rhizobium ruizarguesonis strain SM107 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SING01000006 (7826..7869 [-], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | 21..27, 32..38 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_pSM107_Rh22
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file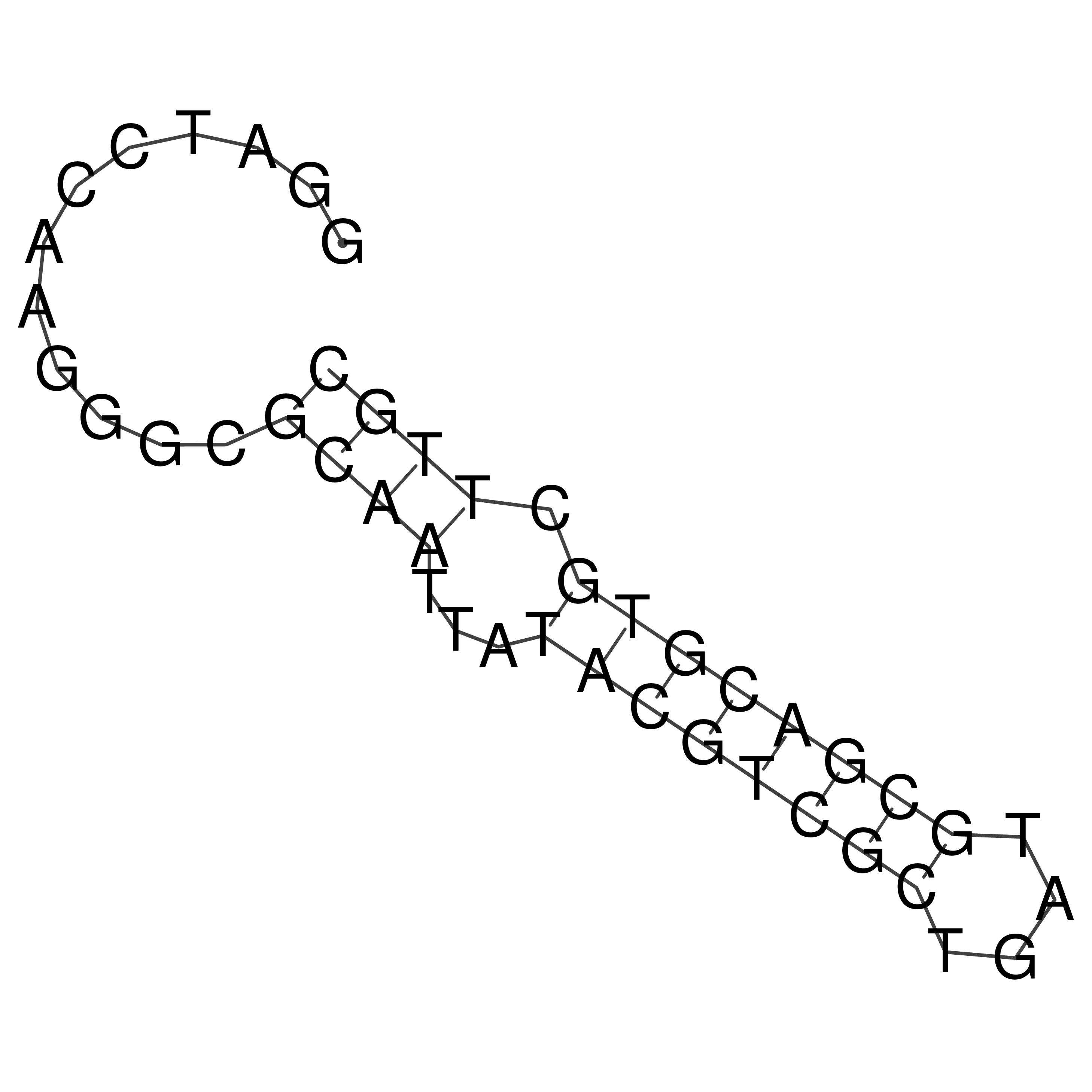
Relaxase
| ID | 1162 | GenBank | WP_130773809 |
| Name | traA_ELH55_RS33975_pSM107_Rh22 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123584.35 Da Isoelectric Point: 9.7853
>WP_130773809.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRALIADR
SVSGAVEAFWNKVEVFEKRSDAQLARDLTIALPLELTPEQNIALVRDFAEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVVGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLVLGG
IDLRIDGRSYEKQGIKLEPTIHLGVGAKAIERKAQERGVRPELERIELNEERRSENTRRILNNPAIVLDL
ITREKSVFDERDIAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKLPARYSTRAMIRLEA
TMVRQALWLSNRDGHAVSEAALDATFRRHERLSQEQKAAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWSRGRDVLDNKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITETRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHAFKTADG
IRQFDAGDQIVFLKNETSLGVKNGMIAHVVEAAANRIVAVVGGGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRRQAKETTLDYEPGKLY
REALRYAENRGLNIVQVARTLLRDQLDWTLRQKTKLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKIFSGSVADTVEQKLDTDPALRKQWEEVSTRFAYVFADPETAFRAINFDAVLVDKETAKLALQTLTTAP
ASIGPLKGKTGILASKTEREERRVADLNVPALKRDLEQYLRMRETAVQRLQADEQGLRQRVSIDIPALSQ
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTLITNAAREPSGKLFDKLSEG
MQPEQKERLTEAWPIMRASQQLAAHERTVQTLRQAEDLRLAPRQTPVMKQ
MAIAHFSASIVSRGGGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRALIADR
SVSGAVEAFWNKVEVFEKRSDAQLARDLTIALPLELTPEQNIALVRDFAEKHILAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVVGEDGQPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLVLGG
IDLRIDGRSYEKQGIKLEPTIHLGVGAKAIERKAQERGVRPELERIELNEERRSENTRRILNNPAIVLDL
ITREKSVFDERDIAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKLPARYSTRAMIRLEA
TMVRQALWLSNRDGHAVSEAALDATFRRHERLSQEQKAAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWSRGRDVLDNKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANVRITETRLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHAFKTADG
IRQFDAGDQIVFLKNETSLGVKNGMIAHVVEAAANRIVAVVGGGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRRQAKETTLDYEPGKLY
REALRYAENRGLNIVQVARTLLRDQLDWTLRQKTKLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMVAG
IKIFSGSVADTVEQKLDTDPALRKQWEEVSTRFAYVFADPETAFRAINFDAVLVDKETAKLALQTLTTAP
ASIGPLKGKTGILASKTEREERRVADLNVPALKRDLEQYLRMRETAVQRLQADEQGLRQRVSIDIPALSQ
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTLITNAAREPSGKLFDKLSEG
MQPEQKERLTEAWPIMRASQQLAAHERTVQTLRQAEDLRLAPRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 745 | GenBank | WP_130773811 |
| Name | traG_ELH55_RS33990_pSM107_Rh22 |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 70349.49 Da Isoelectric Point: 9.5383
>WP_130773811.1 Ti-type conjugative transfer system protein TraG [Rhizobium ruizarguesonis]
MTASKVFLAIVPATTMIVVVVFMPGIEHWLAAFGKTPQAKLILGRIGLALPYATAAAIGTIFLFAANGAA
GIKAAGWGVVSGNGVAILIALLREGVRLAGIAGDVPKGQSVLGYADPATMLGASTAILAGVFALRVAMMG
NAAFAKGSPRRIGGKRAVHGEADWMKIQEAAKLFPDPGGIVVGERYRVDRDSVAAVSFRSGDPSTWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRKAGRKVIILD
PSASGIGFNALDWIGRHGSTKEEDIVAVAIWIMTDNPRSASARDDFFRASAMQLLTALIADVCLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQINRSSGIKGSSRSRSRQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRKDMSASVG
ENRFHKPITEGVKSYKAAPTTDTEAT
MTASKVFLAIVPATTMIVVVVFMPGIEHWLAAFGKTPQAKLILGRIGLALPYATAAAIGTIFLFAANGAA
GIKAAGWGVVSGNGVAILIALLREGVRLAGIAGDVPKGQSVLGYADPATMLGASTAILAGVFALRVAMMG
NAAFAKGSPRRIGGKRAVHGEADWMKIQEAAKLFPDPGGIVVGERYRVDRDSVAAVSFRSGDPSTWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRKAGRKVIILD
PSASGIGFNALDWIGRHGSTKEEDIVAVAIWIMTDNPRSASARDDFFRASAMQLLTALIADVCLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQINRSSGIKGSSRSRSRQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRKDMSASVG
ENRFHKPITEGVKSYKAAPTTDTEAT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 746 | GenBank | WP_130773865 |
| Name | t4cp2_ELH55_RS34335_pSM107_Rh22 |
UniProt ID | _ |
| Length | 821 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 821 a.a. Molecular weight: 91780.57 Da Isoelectric Point: 5.4896
>WP_130773865.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALKSFRHPGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWIIQVEAIRVPTGDYPAEAESYFPDPVTRAIDDERRAHFQTERGHFESRHALILTWRPPEPRRSGLTR
YIYSDAASRSATYADKALENFLTSIREVEQYLANVVSIRRMMTRETSERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDSEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDAVRLQEKCEAIRQLIQAEGFGARIETLNATDAYLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTFGLNGDHYQIGGDIGPSTSGEKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTITPD
YRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGALQCFEIEELM
NMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISD
AERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNMA
LGPVALSFVGATGKEDLKQIRNLHSEHGAAWPLHWLQQRGITHAETLFPAE
MVALKSFRHPGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWIIQVEAIRVPTGDYPAEAESYFPDPVTRAIDDERRAHFQTERGHFESRHALILTWRPPEPRRSGLTR
YIYSDAASRSATYADKALENFLTSIREVEQYLANVVSIRRMMTRETSERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDIPMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDSEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDAVRLQEKCEAIRQLIQAEGFGARIETLNATDAYLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFYPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTFGLNGDHYQIGGDIGPSTSGEKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTITPD
YRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGALQCFEIEELM
NMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISD
AERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNMA
LGPVALSFVGATGKEDLKQIRNLHSEHGAAWPLHWLQQRGITHAETLFPAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 92763..102450
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH55_RS34300 (ELH55_34300) | 88147..89361 | - | 1215 | WP_130773859 | plasmid replication protein RepC | - |
| ELH55_RS34305 (ELH55_34305) | 89516..90502 | - | 987 | WP_130773860 | plasmid partitioning protein RepB | - |
| ELH55_RS34310 (ELH55_34310) | 90577..91764 | - | 1188 | WP_130773861 | plasmid partitioning protein RepA | - |
| ELH55_RS34315 (ELH55_34315) | 92131..92766 | + | 636 | WP_130773862 | acyl-homoserine-lactone synthase | - |
| ELH55_RS34320 (ELH55_34320) | 92763..93734 | + | 972 | WP_130773863 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH55_RS34325 (ELH55_34325) | 93724..94128 | + | 405 | WP_130773864 | conjugal transfer pilin TrbC | virB2 |
| ELH55_RS34330 (ELH55_34330) | 94121..94420 | + | 300 | WP_033184404 | conjugal transfer protein TrbD | virB3 |
| ELH55_RS34335 (ELH55_34335) | 94422..96887 | + | 2466 | WP_130773865 | conjugal transfer protein TrbE | virb4 |
| ELH55_RS34340 (ELH55_34340) | 96859..97668 | + | 810 | WP_130773866 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH55_RS34345 (ELH55_34345) | 97665..97892 | + | 228 | WP_130773867 | entry exclusion protein TrbK | - |
| ELH55_RS34350 (ELH55_34350) | 97886..99076 | + | 1191 | WP_130773868 | P-type conjugative transfer protein TrbL | virB6 |
| ELH55_RS34355 (ELH55_34355) | 99094..99771 | + | 678 | WP_130773869 | conjugal transfer protein TrbF | virB8 |
| ELH55_RS34360 (ELH55_34360) | 99789..100640 | + | 852 | WP_130773870 | P-type conjugative transfer protein TrbG | virB9 |
| ELH55_RS34365 (ELH55_34365) | 100640..101116 | + | 477 | WP_130773871 | conjugal transfer protein TrbH | - |
| ELH55_RS34370 (ELH55_34370) | 101128..102450 | + | 1323 | WP_130773872 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH55_RS34375 (ELH55_34375) | 102580..103680 | - | 1101 | WP_130773873 | LacI family DNA-binding transcriptional regulator | - |
| ELH55_RS34380 (ELH55_34380) | 103826..104848 | + | 1023 | WP_130773874 | SIS domain-containing protein | - |
| ELH55_RS34385 (ELH55_34385) | 104859..105677 | + | 819 | WP_130773875 | PfkB family carbohydrate kinase | - |
| ELH55_RS34390 (ELH55_34390) | 105995..107056 | + | 1062 | WP_130773876 | ABC transporter substrate-binding protein | - |
Host bacterium
| ID | 1609 | GenBank | NZ_SING01000006 |
| Plasmid name | pSM107_Rh22 | Incompatibility group | - |
| Plasmid size | 115807 bp | Coordinate of oriT [Strand] | 7826..7869 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM107 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |