Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101159 |
| Name | oriT_pSS20EcIncFII |
| Organism | Escherichia coli strain EcSS20 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SDDW01000022 (24372..24495 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACACTACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file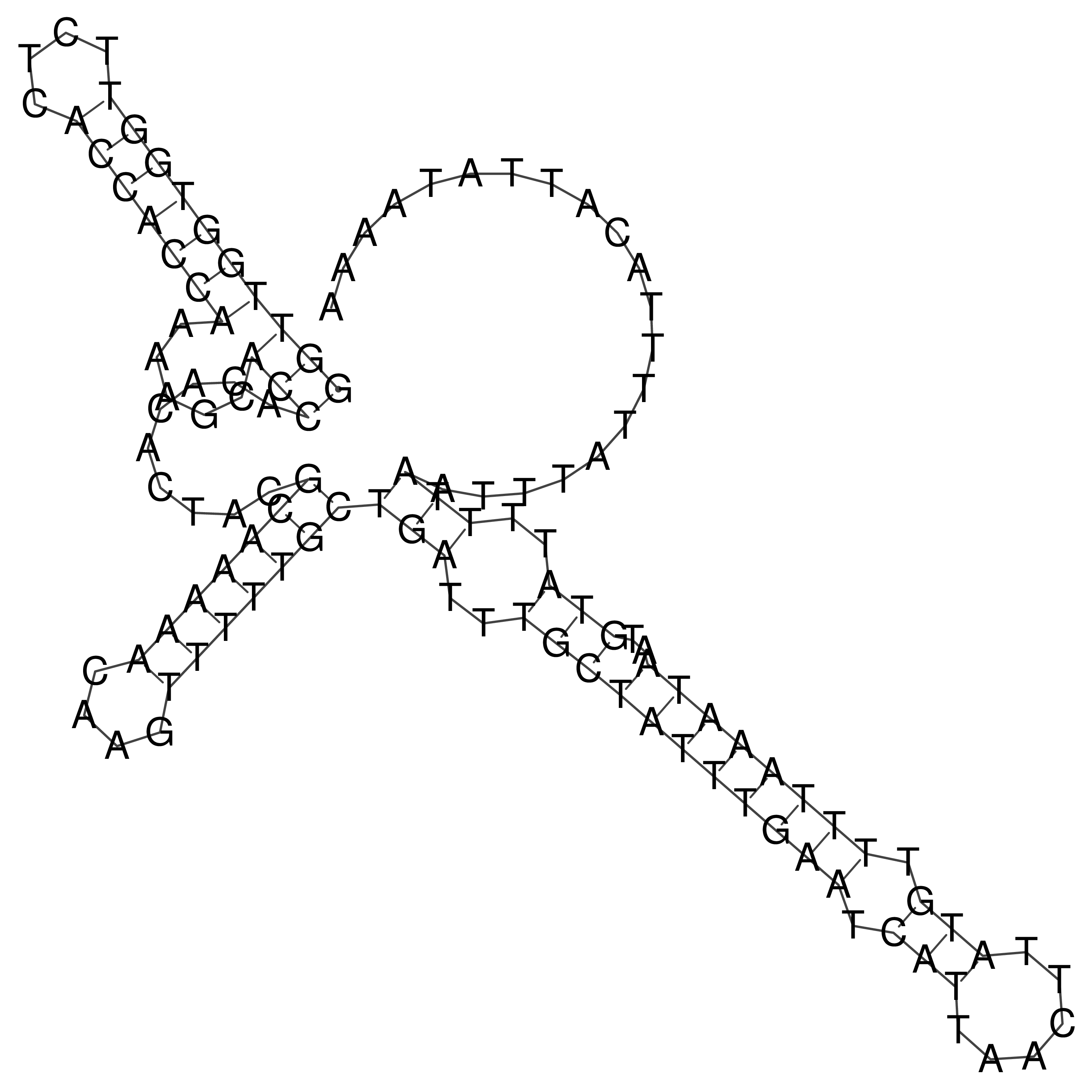
Relaxase
| ID | 1156 | GenBank | WP_129015002 |
| Name | traI_ERL57_RS20135_pSS20EcIncFII |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191766.43 Da Isoelectric Point: 5.7960
MLSFSVVKSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFSENVLANRIAFGKIYQSELRQRVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQDGMTFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQILITDSGQRTGTGSALMSMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISLQKTGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGETFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNMRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRSAISERETALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 415 | GenBank | WP_001151562 |
| Name | WP_001151562_pSS20EcIncFII |
UniProt ID | A0A7I0KNG1 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14420.36 Da Isoelectric Point: 4.8624
MAKVQAYVSDEIVYKINKIVERRRAEGAKSTDVSFSSISTMLLELGLRVHEAQMERKESAFNQAEFNKVL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0KNG1 |
| ID | 416 | GenBank | WP_001254388 |
| Name | WP_001254388_pSS20EcIncFII |
UniProt ID | W9A0Z2 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9072.20 Da Isoelectric Point: 10.2071
MRRRNARGGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVIEENES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W9A0Z2 |
T4CP
| ID | 733 | GenBank | WP_001526033 |
| Name | traC_ERL57_RS20020_pSS20EcIncFII |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99213.73 Da Isoelectric Point: 5.8465
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGINLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASIATQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAGSPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 734 | GenBank | WP_096988748 |
| Name | traD_ERL57_RS20130_pSS20EcIncFII |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 82758.29 Da Isoelectric Point: 5.1008
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILIGLVLWVKISWQTFINGCIYWWCTSLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGHQGKQ
QSENEVTGGRQLTDNPKEVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPEVASGEGVTQAEQPQQPQQPQQPQQ
PQQPQQPVSSVINDKKSDAGVSVPAGGIEQELNMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPD
IQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23800..53511
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ERL57_RS19895 (ERL57_19895) | 20041..20475 | + | 435 | WP_129014999 | conjugation system SOS inhibitor PsiB | - |
| ERL57_RS19900 (ERL57_19900) | 20472..21234 | + | 763 | Protein_28 | plasmid SOS inhibition protein A | - |
| ERL57_RS24610 | 21203..21391 | - | 189 | WP_001336239 | hypothetical protein | - |
| ERL57_RS24990 | 21413..21562 | + | 150 | Protein_30 | plasmid maintenance protein Mok | - |
| ERL57_RS19905 (ERL57_19905) | 21504..21629 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| ERL57_RS24995 (ERL57_19910) | 21849..22079 | + | 231 | WP_071593758 | hypothetical protein | - |
| ERL57_RS19915 (ERL57_19915) | 22077..22274 | - | 198 | Protein_33 | hypothetical protein | - |
| ERL57_RS19920 (ERL57_19920) | 22276..22563 | + | 288 | WP_000107535 | hypothetical protein | - |
| ERL57_RS19925 (ERL57_19925) | 22684..23505 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| ERL57_RS19935 (ERL57_19935) | 23800..24402 | - | 603 | WP_077539848 | transglycosylase SLT domain-containing protein | virB1 |
| ERL57_RS19940 (ERL57_19940) | 24733..25116 | + | 384 | WP_001151562 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| ERL57_RS19945 (ERL57_19945) | 25310..25996 | + | 687 | WP_000332499 | PAS domain-containing protein | - |
| ERL57_RS19950 (ERL57_19950) | 26090..26317 | + | 228 | WP_001254388 | conjugal transfer relaxosome protein TraY | - |
| ERL57_RS19955 (ERL57_19955) | 26351..26710 | + | 360 | WP_016239104 | type IV conjugative transfer system pilin TraA | - |
| ERL57_RS19960 (ERL57_19960) | 26725..27036 | + | 312 | WP_000012113 | type IV conjugative transfer system protein TraL | traL |
| ERL57_RS19965 (ERL57_19965) | 27058..27624 | + | 567 | WP_016239103 | type IV conjugative transfer system protein TraE | traE |
| ERL57_RS19970 (ERL57_19970) | 27611..28339 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| ERL57_RS19975 (ERL57_19975) | 28339..29766 | + | 1428 | WP_129015000 | F-type conjugal transfer pilus assembly protein TraB | traB |
| ERL57_RS19980 (ERL57_19980) | 29756..30346 | + | 591 | WP_001535745 | conjugal transfer pilus-stabilizing protein TraP | - |
| ERL57_RS19985 (ERL57_19985) | 30333..30653 | + | 321 | WP_001057298 | conjugal transfer protein TrbD | virb4 |
| ERL57_RS19990 (ERL57_19990) | 30646..30897 | + | 252 | WP_001038333 | conjugal transfer protein TrbG | - |
| ERL57_RS19995 (ERL57_19995) | 30894..31409 | + | 516 | WP_032234029 | type IV conjugative transfer system lipoprotein TraV | traV |
| ERL57_RS20000 (ERL57_20000) | 31544..31765 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| ERL57_RS20005 (ERL57_20005) | 31758..32234 | + | 477 | WP_109547838 | hypothetical protein | - |
| ERL57_RS20010 (ERL57_20010) | 32315..32533 | + | 219 | WP_000556751 | hypothetical protein | - |
| ERL57_RS20015 (ERL57_20015) | 32561..32908 | + | 348 | WP_000836682 | hypothetical protein | - |
| ERL57_RS20020 (ERL57_20020) | 33034..35664 | + | 2631 | WP_001526033 | type IV secretion system protein TraC | virb4 |
| ERL57_RS20025 (ERL57_20025) | 35661..36047 | + | 387 | WP_000214084 | type-F conjugative transfer system protein TrbI | - |
| ERL57_RS20030 (ERL57_20030) | 36044..36676 | + | 633 | WP_001526032 | type-F conjugative transfer system protein TraW | traW |
| ERL57_RS20035 (ERL57_20035) | 36673..37665 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| ERL57_RS20040 (ERL57_20040) | 37692..38000 | + | 309 | WP_000412445 | hypothetical protein | - |
| ERL57_RS20045 (ERL57_20045) | 38063..38581 | + | 519 | WP_000009085 | hypothetical protein | - |
| ERL57_RS20050 (ERL57_20050) | 38608..39246 | + | 639 | WP_109547839 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| ERL57_RS20055 (ERL57_20055) | 39243..39614 | + | 372 | WP_000056340 | hypothetical protein | - |
| ERL57_RS20060 (ERL57_20060) | 39639..40061 | + | 423 | WP_001229306 | HNH endonuclease signature motif containing protein | - |
| ERL57_RS20065 (ERL57_20065) | 40058..41908 | + | 1851 | WP_109547840 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| ERL57_RS20070 (ERL57_20070) | 41935..42192 | + | 258 | WP_129015006 | conjugal transfer protein TrbE | - |
| ERL57_RS20075 (ERL57_20075) | 42185..42928 | + | 744 | WP_001030385 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| ERL57_RS20080 (ERL57_20080) | 42942..43286 | + | 345 | WP_096979571 | conjugal transfer protein TrbA | - |
| ERL57_RS20085 (ERL57_20085) | 43405..43689 | + | 285 | WP_000624194 | type-F conjugative transfer system pilin chaperone TraQ | - |
| ERL57_RS20090 (ERL57_20090) | 43676..44221 | + | 546 | WP_129015001 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| ERL57_RS20095 (ERL57_20095) | 44151..44498 | + | 348 | WP_001309242 | P-type conjugative transfer protein TrbJ | - |
| ERL57_RS20100 (ERL57_20100) | 44479..44871 | + | 393 | WP_000660700 | F-type conjugal transfer protein TrbF | - |
| ERL57_RS20105 (ERL57_20105) | 44858..46231 | + | 1374 | WP_021563037 | conjugal transfer pilus assembly protein TraH | traH |
| ERL57_RS20110 (ERL57_20110) | 46228..49050 | + | 2823 | WP_109547842 | conjugal transfer mating-pair stabilization protein TraG | traG |
| ERL57_RS20115 (ERL57_20115) | 49066..49551 | + | 486 | WP_020241360 | surface exclusion inner membrane protein | - |
| ERL57_RS20120 (ERL57_20120) | 49600..50331 | + | 732 | WP_000782451 | conjugal transfer complement resistance protein TraT | - |
| ERL57_RS20125 (ERL57_20125) | 50534..51271 | + | 738 | WP_032301641 | hypothetical protein | - |
| ERL57_RS20130 (ERL57_20130) | 51322..53511 | + | 2190 | WP_096988748 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 1603 | GenBank | NZ_SDDW01000022 |
| Plasmid name | pSS20EcIncFII | Incompatibility group | IncFII |
| Plasmid size | 66445 bp | Coordinate of oriT [Strand] | 24372..24495 [+] |
| Host baterium | Escherichia coli strain EcSS20 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |