Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101157 |
| Name | oriT_pSM157B_Rh08 |
| Organism | Rhizobium ruizarguesonis strain SM157B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQH01000006 (91189..91222 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM157B_Rh08
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file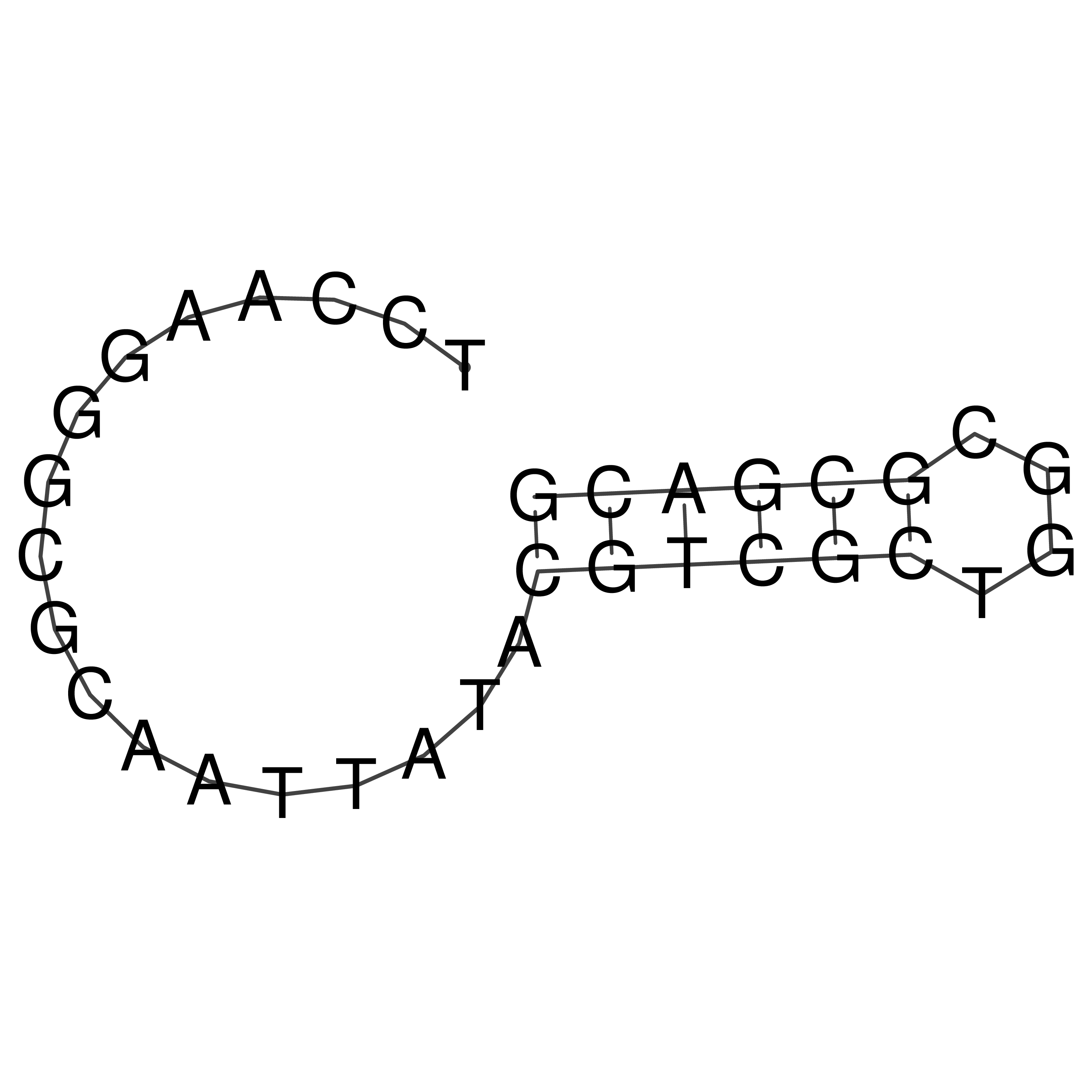
Relaxase
| ID | 1154 | GenBank | WP_130657460 |
| Name | traA_ELI34_RS29470_pSM157B_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122987.83 Da Isoelectric Point: 9.6219
>WP_130657460.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 729 | GenBank | WP_065276635 |
| Name | t4cp2_ELI34_RS29335_pSM157B_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91588.38 Da Isoelectric Point: 5.9977
>WP_065276635.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 730 | GenBank | WP_130657461 |
| Name | traG_ELI34_RS29485_pSM157B_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69518.76 Da Isoelectric Point: 8.9386
>WP_130657461.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 62699..72210
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI34_RS29300 (ELI34_29305) | 58026..59240 | - | 1215 | WP_130666985 | plasmid replication protein RepC | - |
| ELI34_RS29305 (ELI34_29310) | 59430..60467 | - | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELI34_RS29310 (ELI34_29315) | 60464..61687 | - | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELI34_RS29315 (ELI34_29320) | 62064..62702 | + | 639 | WP_130657452 | acyl-homoserine-lactone synthase | - |
| ELI34_RS29320 (ELI34_29325) | 62699..63676 | + | 978 | WP_065276632 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI34_RS29325 (ELI34_29330) | 63666..64049 | + | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELI34_RS29330 (ELI34_29335) | 64042..64341 | + | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELI34_RS29335 (ELI34_29340) | 64352..66802 | + | 2451 | WP_065276635 | conjugal transfer protein TrbE | virb4 |
| ELI34_RS29340 (ELI34_29345) | 66780..67577 | + | 798 | WP_130657453 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI34_RS29345 (ELI34_29350) | 67574..67774 | + | 201 | WP_065276637 | entry exclusion protein TrbK | - |
| ELI34_RS29350 (ELI34_29355) | 67768..68949 | + | 1182 | WP_065276638 | P-type conjugative transfer protein TrbL | virB6 |
| ELI34_RS29355 (ELI34_29360) | 68970..69632 | + | 663 | WP_065276639 | conjugal transfer protein TrbF | virB8 |
| ELI34_RS29360 (ELI34_29365) | 69650..70462 | + | 813 | WP_065276640 | P-type conjugative transfer protein TrbG | virB9 |
| ELI34_RS29365 (ELI34_29370) | 70466..70900 | + | 435 | WP_130657454 | conjugal transfer protein TrbH | - |
| ELI34_RS29370 (ELI34_29375) | 70912..72210 | + | 1299 | WP_130660093 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI34_RS29375 (ELI34_29380) | 72656..72997 | + | 342 | WP_063963361 | transposase | - |
| ELI34_RS29380 (ELI34_29385) | 72994..73341 | + | 348 | WP_057210537 | IS66 family insertion sequence element accessory protein TnpB | - |
| ELI34_RS29385 (ELI34_29390) | 73420..75078 | + | 1659 | WP_130654681 | IS66 family transposase | - |
| ELI34_RS29390 (ELI34_29395) | 75078..75662 | + | 585 | WP_130654682 | plasmid pRiA4b ORF-3 family protein | - |
| ELI34_RS29395 (ELI34_29400) | 75710..76357 | + | 648 | WP_130666986 | autoinducer binding domain-containing protein | - |
| ELI34_RS29400 (ELI34_29405) | 76361..76684 | - | 324 | WP_130657456 | transcriptional repressor TraM | - |
| ELI34_RS29405 (ELI34_29410) | 76907..77149 | + | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
Host bacterium
| ID | 1601 | GenBank | NZ_SIQH01000006 |
| Plasmid name | pSM157B_Rh08 | Incompatibility group | - |
| Plasmid size | 280083 bp | Coordinate of oriT [Strand] | 91189..91222 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM157B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, fixG, fixQ, fixO, fixN, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifT, nodJ, nodI, nodC, nodA, nodF, nodE, nodM |
| Anti-CRISPR | - |