Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101155 |
| Name | oriT_pSM143_Rh07 |
| Organism | Rhizobium ruizarguesonis strain SM143 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOZ01000007 (29375..29415 [-], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM143_Rh07
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file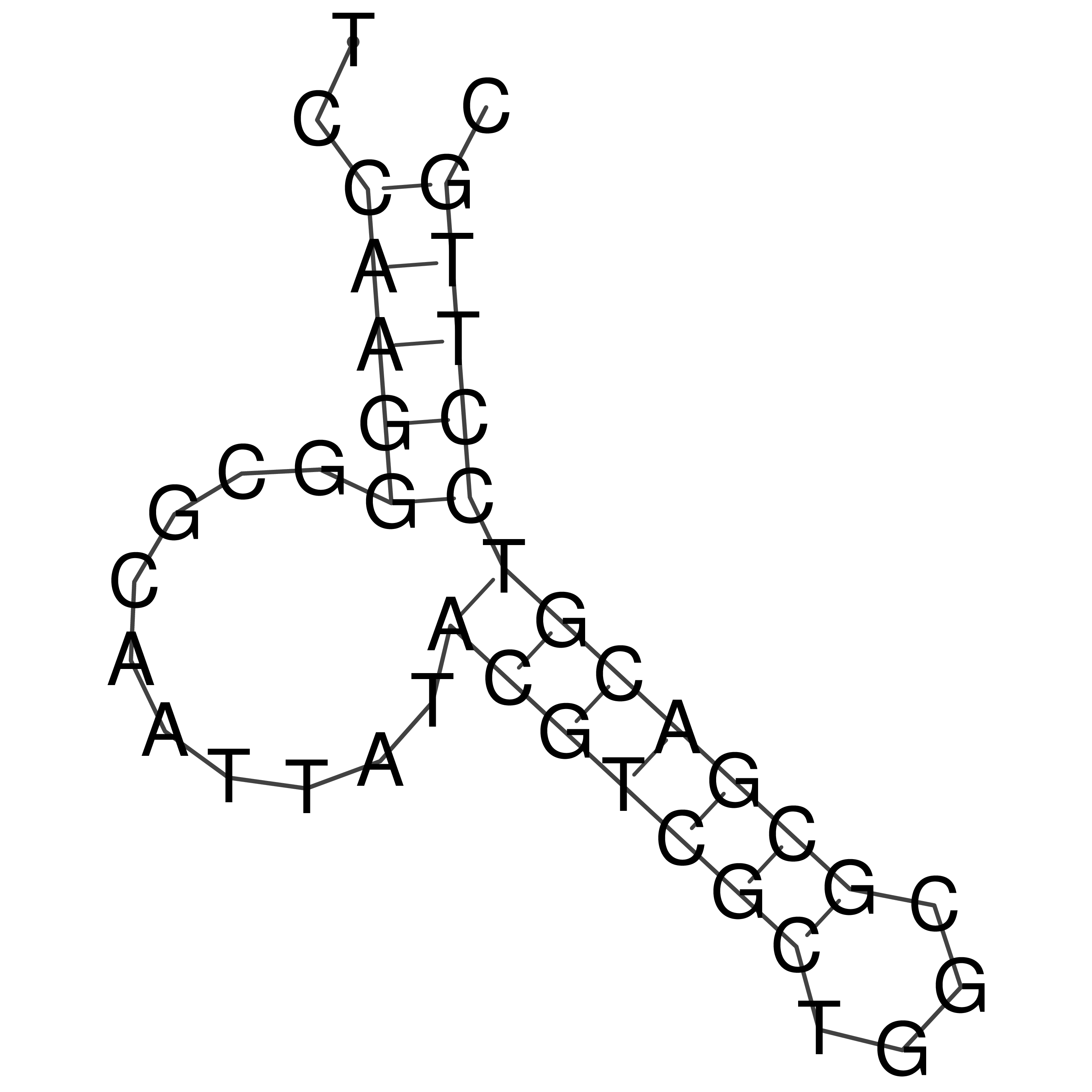
Relaxase
| ID | 1152 | GenBank | WP_130692296 |
| Name | traA_ELI00_RS33040_pSM143_Rh07 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122959.16 Da Isoelectric Point: 9.4990
>WP_130692296.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRQVTIEQRFYNNLDHGYAT
TIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEK
SALCRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKALGLVRGSNKHSIPDTIK
EARPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVEDQSVAQST
LARIAGEPESFGALKGKTGVLASRGDKHDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKVS
VDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDGK
TFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADS
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDDKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVLLFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
VRLEAEMANRAIWLSGRASHGVREEVLQATFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRVGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAAGGIIAEIGEGEHRQVTIEQRFYNNLDHGYAT
TIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEK
SALCRQAFRFAEARGLHLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGKALGLVRGSNKHSIPDTIK
EARPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAAFKAINVDAMVEDQSVAQST
LARIAGEPESFGALKGKTGVLASRGDKHDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKVS
VDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDGK
TFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 725 | GenBank | WP_130692276 |
| Name | t4cp2_ELI00_RS32955_pSM143_Rh07 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91738.38 Da Isoelectric Point: 5.7923
>WP_130692276.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLKYV
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLKYV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 726 | GenBank | WP_130692298 |
| Name | traG_ELI00_RS33055_pSM143_Rh07 |
UniProt ID | _ |
| Length | 643 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 643 a.a. Molecular weight: 69483.71 Da Isoelectric Point: 9.1051
>WP_130692298.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDEK
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPRPA
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSAFMTVPADKTVWAFLDPATSIGASAALLCACFALRVALIGN
AAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGGK
SPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLDP
RKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDEK
DQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAA
LVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGYM
RIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTV
EIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGT
NRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9962..31989
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI00_RS32915 (ELI00_32915) | 5492..6514 | - | 1023 | WP_128408794 | plasmid partitioning protein RepB | - |
| ELI00_RS32920 (ELI00_32920) | 6511..7725 | - | 1215 | WP_027691186 | plasmid partitioning protein RepA | - |
| ELI00_RS32925 (ELI00_32925) | 8342..8596 | - | 255 | WP_130692270 | IS66 family insertion sequence element accessory protein TnpB | - |
| ELI00_RS39965 | 8637..8849 | - | 213 | WP_245458276 | hypothetical protein | - |
| ELI00_RS32935 (ELI00_32935) | 9327..9965 | + | 639 | WP_130692272 | acyl-homoserine-lactone synthase | - |
| ELI00_RS32940 (ELI00_32940) | 9962..10939 | + | 978 | WP_130692274 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI00_RS32945 (ELI00_32945) | 10929..11312 | + | 384 | WP_063473648 | TrbC/VirB2 family protein | virB2 |
| ELI00_RS32950 (ELI00_32950) | 11305..11604 | + | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELI00_RS32955 (ELI00_32955) | 11615..14071 | + | 2457 | WP_130692276 | conjugal transfer protein TrbE | virb4 |
| ELI00_RS32960 (ELI00_32960) | 14043..14846 | + | 804 | WP_128408799 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI00_RS32965 (ELI00_32965) | 14843..15043 | + | 201 | WP_128408800 | entry exclusion protein TrbK | - |
| ELI00_RS32970 (ELI00_32970) | 15037..16218 | + | 1182 | WP_130692278 | P-type conjugative transfer protein TrbL | virB6 |
| ELI00_RS32975 (ELI00_32975) | 16239..16901 | + | 663 | WP_130692280 | conjugal transfer protein TrbF | virB8 |
| ELI00_RS32980 (ELI00_32980) | 16919..17731 | + | 813 | WP_130692282 | P-type conjugative transfer protein TrbG | virB9 |
| ELI00_RS32985 (ELI00_32985) | 17735..18175 | + | 441 | WP_130692284 | conjugal transfer protein TrbH | - |
| ELI00_RS32990 (ELI00_32990) | 18187..19485 | + | 1299 | WP_130692286 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI00_RS32995 (ELI00_32995) | 19794..20498 | + | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| ELI00_RS33000 (ELI00_33000) | 20483..20824 | - | 342 | WP_128408807 | transcriptional repressor TraM | - |
| ELI00_RS33005 (ELI00_33005) | 21050..21292 | + | 243 | WP_128408808 | helix-turn-helix transcriptional regulator | - |
| ELI00_RS33010 (ELI00_33010) | 21406..22038 | + | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| ELI00_RS33015 (ELI00_33015) | 22035..22409 | + | 375 | WP_130692288 | DUF5615 family PIN-like protein | - |
| ELI00_RS33020 (ELI00_33020) | 22766..23481 | + | 716 | Protein_25 | Crp/Fnr family transcriptional regulator | - |
| ELI00_RS33025 (ELI00_33025) | 23626..24246 | - | 621 | WP_130692290 | TraH family protein | virB1 |
| ELI00_RS33030 (ELI00_33030) | 24264..25430 | - | 1167 | WP_130692292 | conjugal transfer protein TraB | - |
| ELI00_RS33035 (ELI00_33035) | 25420..25983 | - | 564 | WP_130692294 | conjugative transfer signal peptidase TraF | - |
| ELI00_RS33040 (ELI00_33040) | 25980..29303 | - | 3324 | WP_130692296 | Ti-type conjugative transfer relaxase TraA | - |
| ELI00_RS33045 (ELI00_33045) | 29558..29851 | + | 294 | WP_128409207 | conjugal transfer protein TraC | - |
| ELI00_RS33050 (ELI00_33050) | 29856..30071 | + | 216 | WP_011654561 | type IV conjugative transfer system coupling protein TraD | - |
| ELI00_RS33055 (ELI00_33055) | 30058..31989 | + | 1932 | WP_130692298 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELI00_RS33060 (ELI00_33060) | 32427..32876 | + | 450 | WP_130692300 | thermonuclease family protein | - |
| ELI00_RS33065 (ELI00_33065) | 33118..33432 | - | 315 | WP_130692302 | hypothetical protein | - |
| ELI00_RS33070 (ELI00_33070) | 34213..35706 | + | 1494 | WP_130764305 | group II intron reverse transcriptase/maturase | - |
| ELI00_RS33075 (ELI00_33075) | 36058..36366 | + | 309 | WP_063473655 | WGR domain-containing protein | - |
| ELI00_RS39325 | 36434..36607 | - | 174 | WP_165403408 | hypothetical protein | - |
Host bacterium
| ID | 1599 | GenBank | NZ_SIOZ01000007 |
| Plasmid name | pSM143_Rh07 | Incompatibility group | - |
| Plasmid size | 62178 bp | Coordinate of oriT [Strand] | 29375..29415 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM143 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD |
| Anti-CRISPR | - |