Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101142 |
| Name | oriT_pSM157C_Rh03_Rh08 |
| Organism | Rhizobium ruizarguesonis strain SM157C |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQI01000004 (34193..34226 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM157C_Rh03_Rh08
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file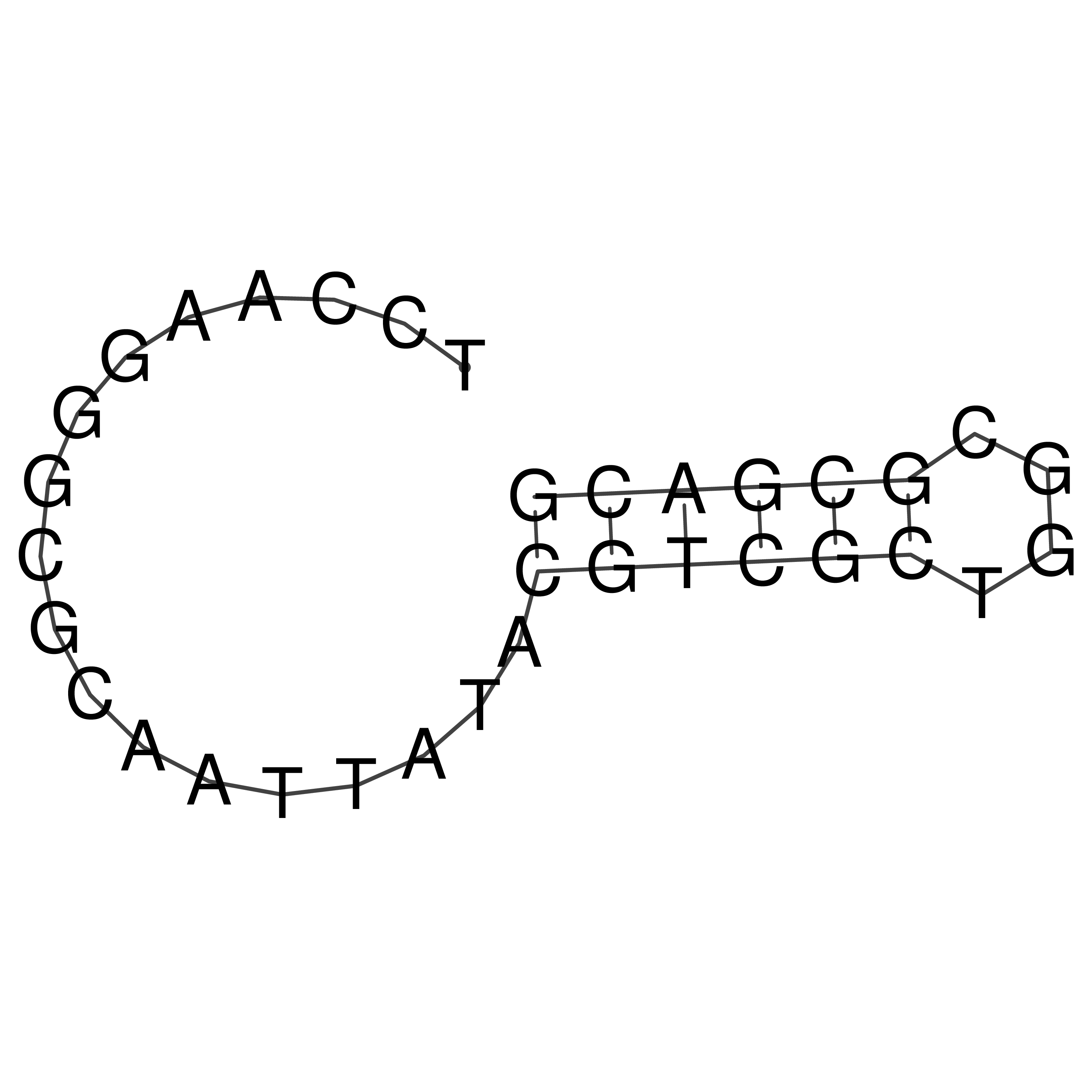
Relaxase
| ID | 1141 | GenBank | WP_130680553 |
| Name | traA_ELI35_RS30210_pSM157C_Rh03_Rh08 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122693.44 Da Isoelectric Point: 9.6904
>WP_130680553.1 Ti-type conjugative transfer relaxase TraA [Rhizobium ruizarguesonis]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTGG
KTFETVTAGMTAGQKAEVQSAWNAMRTVQQLGSHERTTEAHKQAETLQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTGG
KTFETVTAGMTAGQKAEVQSAWNAMRTVQQLGSHERTTEAHKQAETLQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 703 | GenBank | WP_130680549 |
| Name | t4cp2_ELI35_RS30095_pSM157C_Rh03_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91492.16 Da Isoelectric Point: 5.6886
>WP_130680549.1 conjugal transfer protein TrbE [Rhizobium ruizarguesonis]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREVLQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAANSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGNSLLPLTLAASGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVAITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREVLQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAANSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGNSLLPLTLAASGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVAITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 704 | GenBank | WP_130657461 |
| Name | traG_ELI35_RS30225_pSM157C_Rh03_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69518.76 Da Isoelectric Point: 8.9386
>WP_130657461.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSVHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDGGGVRGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKVRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8837..18343
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI35_RS30060 (ELI35_30070) | 4164..5378 | - | 1215 | WP_130666985 | plasmid replication protein RepC | - |
| ELI35_RS30065 (ELI35_30075) | 5568..6605 | - | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELI35_RS30070 (ELI35_30080) | 6602..7825 | - | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELI35_RS30075 (ELI35_30085) | 8202..8840 | + | 639 | WP_130657452 | acyl-homoserine-lactone synthase | - |
| ELI35_RS30080 (ELI35_30090) | 8837..9814 | + | 978 | WP_130680548 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI35_RS30085 (ELI35_30095) | 9804..10187 | + | 384 | WP_130680522 | TrbC/VirB2 family protein | virB2 |
| ELI35_RS30090 (ELI35_30100) | 10180..10479 | + | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELI35_RS30095 (ELI35_30105) | 10490..12940 | + | 2451 | WP_130680549 | conjugal transfer protein TrbE | virb4 |
| ELI35_RS30100 (ELI35_30110) | 12918..13715 | + | 798 | WP_130657453 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI35_RS30105 (ELI35_30115) | 13712..13912 | + | 201 | WP_065276637 | entry exclusion protein TrbK | - |
| ELI35_RS30110 (ELI35_30120) | 13906..15087 | + | 1182 | WP_065276638 | P-type conjugative transfer protein TrbL | virB6 |
| ELI35_RS30115 (ELI35_30125) | 15108..15765 | + | 658 | Protein_15 | conjugal transfer protein TrbF | - |
| ELI35_RS30120 (ELI35_30130) | 15783..16595 | + | 813 | WP_065276640 | P-type conjugative transfer protein TrbG | virB9 |
| ELI35_RS30125 (ELI35_30135) | 16599..17033 | + | 435 | WP_130657454 | conjugal transfer protein TrbH | - |
| ELI35_RS30130 (ELI35_30140) | 17045..18343 | + | 1299 | WP_130660093 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI35_RS30135 (ELI35_30145) | 18654..19358 | + | 705 | WP_130654288 | autoinducer binding domain-containing protein | - |
| ELI35_RS30140 (ELI35_30150) | 19362..19685 | - | 324 | WP_130657456 | transcriptional repressor TraM | - |
| ELI35_RS30145 (ELI35_30155) | 19908..20150 | + | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
| ELI35_RS40230 (ELI35_30160) | 20253..20690 | + | 438 | Protein_22 | hypothetical protein | - |
| ELI35_RS30155 (ELI35_30165) | 20781..21398 | + | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELI35_RS30160 (ELI35_30170) | 21391..22260 | + | 870 | Protein_24 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELI35_RS30165 (ELI35_30175) | 22471..22881 | + | 411 | WP_018244910 | transposase | - |
| ELI35_RS30170 (ELI35_30180) | 22878..23123 | + | 246 | WP_130660511 | IS66 family insertion sequence element accessory protein TnpB | - |
Host bacterium
| ID | 1586 | GenBank | NZ_SIQI01000004 |
| Plasmid name | pSM157C_Rh03_Rh08 | Incompatibility group | - |
| Plasmid size | 280831 bp | Coordinate of oriT [Strand] | 34193..34226 [-] |
| Host baterium | Rhizobium ruizarguesonis strain SM157C |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, fixG, fixQ, fixO, fixN, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifT, nodJ, nodI, nodC, nodA, nodF, nodE, nodM |
| Anti-CRISPR | - |