Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101090 |
| Name | oriT_pZJ71 |
| Organism | Escherichia coli strain ZJ71 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MCAM01000040 (43507..43559 [-], 53 nt) |
| oriT length | 53 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 53 nt
>oriT_pZJ71
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
CACACGATTGTAACATGACCGGAACGGTCTTGTGTACAATCGGTATCGTGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file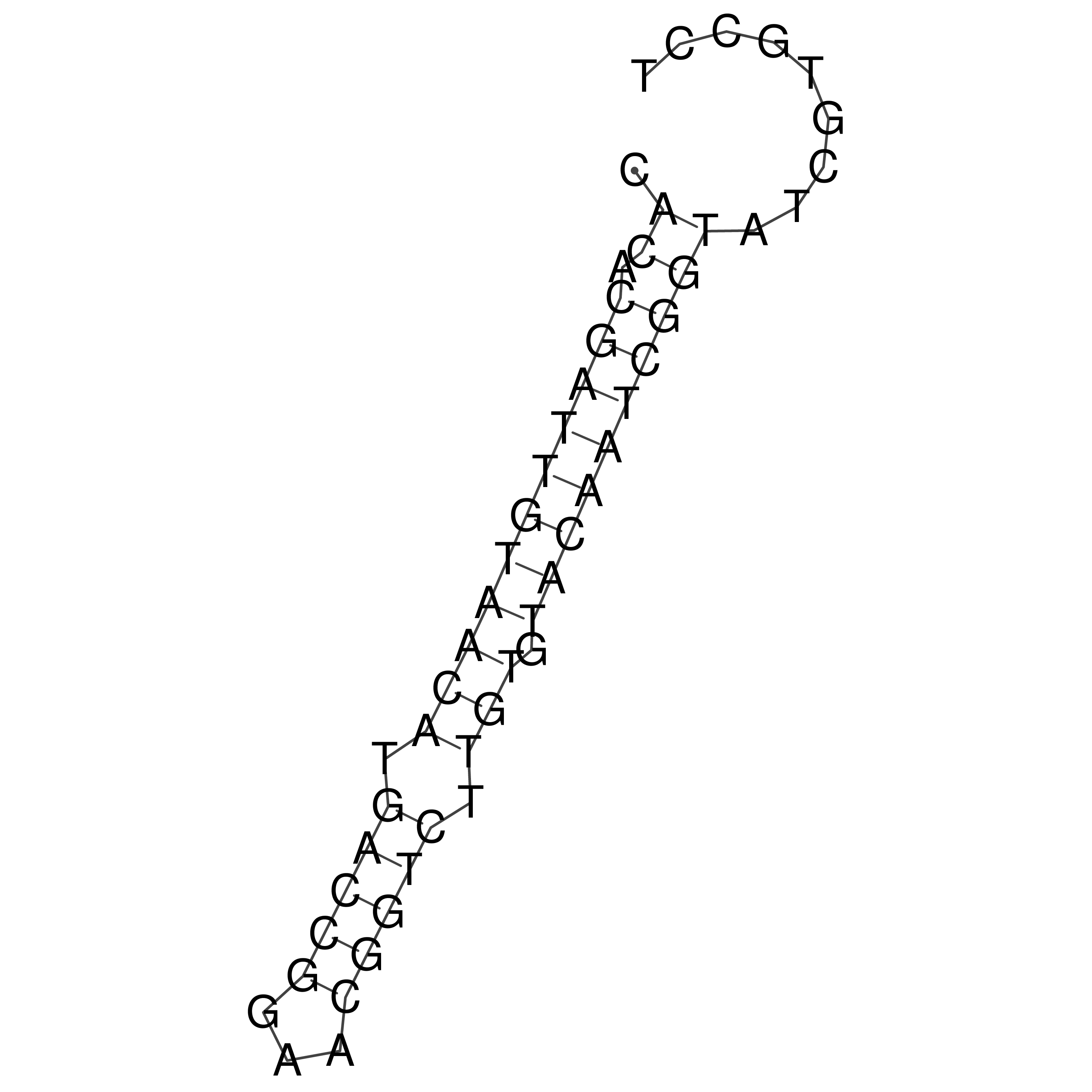
T4CP
| ID | 673 | GenBank | WP_015059539 |
| Name | t4cp2_BED25_RS22345_pZJ71 |
UniProt ID | _ |
| Length | 652 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 652 a.a. Molecular weight: 73404.02 Da Isoelectric Point: 9.4339
>WP_015059539.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacteriaceae]
MNAKKMGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
MNAKKMGGLILFLLLLLVGVLIASNYLGGYTALRYSSVDMSLLKWDTFHSVISTFSGNPQYKKLVFMAWF
GFSVPLIFFAIFMLIVVIGIMPKKVIYGDARLATDMDLSKSGFFPDKKSPYKHPPILIGKMFKGRYKKQF
IYFAGQQFLILYAPTRSGKGVGIVIPNCVNYPGSMVILDIKLENWFLSAGFRQKELGQKCFLFAPAGYAE
TIDQAIKGQIRSHRWNPLDCVSRSDLLRETDLAKIAAILIPASDDPIWSDSARNLFVGLGLYLLDKERFH
LDQKAKGHNAPDVLVSISAILKTSIPDNGKDLAAWMGQEVENRSWISDKTKSFFFEFMSAPDRTRGSIKT
NFSSPLNIFSNPVTAEATNFSDFDIRDIRKKPMSIYLGLTPDALITHEKIVNLFFSLLVNENCRELPEHN
PDLKYQCLILLDEFTSMGKSEVIERAVGFTAGYNLRFMFILQNEGQGQKSDMYGQEGWTTFTENSAVVLY
YPPKSKNALAKKISEEIGVRDMKISKRSISSGGGKGGSSRTRNDDVIERPVLLPEEIVSLRDKKNKARNI
AIREIITSEFSRPFIANKIIWFEEPEFKRRVDIARNNHVDIPNLFTQEVMDEIAKIAEIYLPKAGGKKVM
VAGGNVITNPDLDNHDKTDVSE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1687..25144
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BED25_RS22295 (BED25_24280) | 1..1035 | - | 1035 | WP_000750512 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| BED25_RS22300 (BED25_24285) | 1048..1683 | - | 636 | WP_000934977 | A24 family peptidase | - |
| BED25_RS22305 (BED25_24290) | 1687..2169 | - | 483 | WP_001258095 | lytic transglycosylase domain-containing protein | virB1 |
| BED25_RS22310 (BED25_24295) | 2235..2792 | - | 558 | WP_000095048 | type 4 pilus major pilin | - |
| BED25_RS22315 (BED25_24300) | 2837..3946 | - | 1110 | WP_000974903 | type II secretion system F family protein | - |
| BED25_RS22320 (BED25_24305) | 3937..5475 | - | 1539 | WP_000466225 | ATPase, T2SS/T4P/T4SS family | virB11 |
| BED25_RS22325 (BED25_24310) | 5500..5994 | - | 495 | WP_000912553 | type IV pilus biogenesis protein PilP | - |
| BED25_RS22330 (BED25_24315) | 5978..7288 | - | 1311 | WP_001454111 | type 4b pilus protein PilO2 | - |
| BED25_RS22335 (BED25_24320) | 7339..8982 | - | 1644 | WP_001035592 | PilN family type IVB pilus formation outer membrane protein | - |
| BED25_RS22340 (BED25_24325) | 8975..9511 | - | 537 | WP_001220543 | sigma 54-interacting transcriptional regulator | virb4 |
| BED25_RS22345 (BED25_24330) | 9558..11516 | - | 1959 | WP_015059539 | type IV secretory system conjugative DNA transfer family protein | - |
| BED25_RS22350 (BED25_24335) | 11532..12587 | - | 1056 | WP_001059977 | P-type DNA transfer ATPase VirB11 | virB11 |
| BED25_RS22355 (BED25_24340) | 12606..13745 | - | 1140 | WP_171264649 | TrbI/VirB10 family protein | virB10 |
| BED25_RS22360 (BED25_24345) | 13735..14436 | - | 702 | WP_000274524 | TrbG/VirB9 family P-type conjugative transfer protein | - |
| BED25_RS22365 (BED25_24350) | 14502..15236 | - | 735 | WP_000432282 | type IV secretion system protein | virB8 |
| BED25_RS22370 (BED25_24360) | 15402..17759 | - | 2358 | WP_063078682 | VirB4 family type IV secretion system protein | virb4 |
| BED25_RS22375 (BED25_24365) | 17765..18085 | - | 321 | WP_000362080 | VirB3 family type IV secretion system protein | virB3 |
| BED25_RS22380 (BED25_24370) | 18156..18446 | - | 291 | WP_000865479 | conjugal transfer protein | - |
| BED25_RS22385 (BED25_24375) | 18446..19030 | - | 585 | WP_001177113 | lytic transglycosylase domain-containing protein | virB1 |
| BED25_RS22390 (BED25_24380) | 19051..19449 | - | 399 | WP_001153665 | hypothetical protein | - |
| BED25_RS22395 (BED25_24385) | 19568..20005 | - | 438 | WP_000539665 | type IV pilus biogenesis protein PilM | - |
| BED25_RS22400 (BED25_24390) | 20011..21246 | - | 1236 | WP_015059538 | TcpQ domain-containing protein | - |
| BED25_RS22405 (BED25_24395) | 21249..21548 | - | 300 | WP_000835764 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| BED25_RS22410 (BED25_24400) | 21616..21897 | - | 282 | WP_000638823 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| BED25_RS22415 (BED25_24405) | 21887..22138 | - | 252 | WP_000121741 | hypothetical protein | - |
| BED25_RS22420 (BED25_24410) | 22238..22873 | - | 636 | WP_015059536 | hypothetical protein | - |
| BED25_RS22425 (BED25_24415) | 22946..23233 | - | 288 | WP_001032611 | EexN family lipoprotein | - |
| BED25_RS22430 (BED25_24420) | 23246..23500 | - | 255 | WP_001043555 | EexN family lipoprotein | - |
| BED25_RS22435 (BED25_24425) | 23502..24143 | - | 642 | WP_001425343 | type IV secretion system protein | - |
| BED25_RS22440 (BED25_24430) | 24149..25144 | - | 996 | WP_001028543 | type IV secretion system protein | virB6 |
| BED25_RS22445 (BED25_24435) | 25148..25405 | - | 258 | WP_000739144 | hypothetical protein | - |
| BED25_RS22450 | 25402..25704 | - | 303 | WP_001360345 | hypothetical protein | - |
| BED25_RS22455 (BED25_24440) | 25685..25942 | - | 258 | WP_001542015 | hypothetical protein | - |
| BED25_RS22460 (BED25_24445) | 25975..26421 | - | 447 | WP_001243165 | hypothetical protein | - |
| BED25_RS22465 | 26432..26602 | - | 171 | WP_000550720 | hypothetical protein | - |
| BED25_RS22470 | 26606..27049 | - | 444 | WP_000964330 | NfeD family protein | - |
| BED25_RS22475 (BED25_24455) | 27423..28376 | - | 954 | WP_072089442 | SPFH domain-containing protein | - |
| BED25_RS22480 | 28403..28579 | - | 177 | WP_000753050 | hypothetical protein | - |
| BED25_RS22485 (BED25_24460) | 28572..28787 | - | 216 | WP_001127357 | DUF1187 family protein | - |
| BED25_RS22490 (BED25_24465) | 28780..29232 | - | 453 | WP_000101552 | CaiF/GrlA family transcriptional regulator | - |
Host bacterium
| ID | 1534 | GenBank | NZ_MCAM01000040 |
| Plasmid name | pZJ71 | Incompatibility group | IncI2 |
| Plasmid size | 58964 bp | Coordinate of oriT [Strand] | 43507..43559 [-] |
| Host baterium | Escherichia coli strain ZJ71 |
Cargo genes
| Drug resistance gene | mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |