Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101057 |
| Name | oriT_pUnk_2670 |
| Organism | Escherichia coli strain PH-2670-18 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_WLVN01000005 (95065..95150 [+], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
AATTACATGATTTAAAACACAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file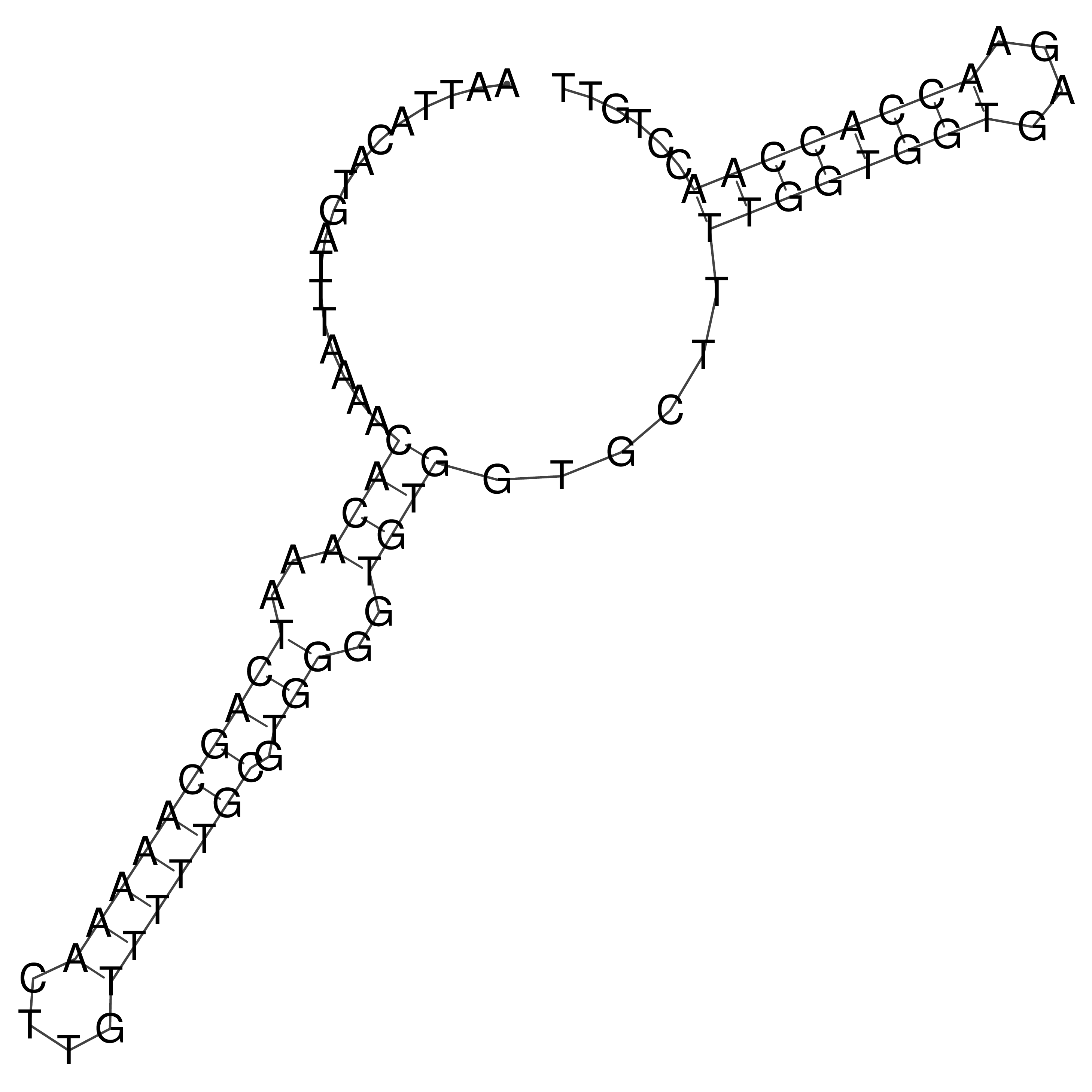
Relaxase
| ID | 1104 | GenBank | WP_021518366 |
| Name | traI_GKM64_RS20795_pUnk_2670 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192172.86 Da Isoelectric Point: 6.1609
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIRETVGEDASLKSRDVAALDTRKSKQHVDPEIKMTEWMQT
LKETGFDIRAYRDAAEQRAYTRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
AGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVTSVSEDAMTVVVPGRAEPATLPVSDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFAAEGTSFTELGREIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVEKALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAQRLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRTATSERETALPESVLRESQREREAVREVARENLLQRLLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 400 | GenBank | WP_001352842 |
| Name | WP_001352842_pUnk_2670 |
UniProt ID | A0A6Y3H4U0 |
| Length | 71 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 71 a.a. Molecular weight: 8117.19 Da Isoelectric Point: 9.9216
MSRGRTRAAPGNKVLLILDETTNQKLLAARDRSGRTKTNEVFIRLKDHLNRFPDFYNASLVKEEAESIDD
I
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A6Y3H4U0 |
| ID | 401 | GenBank | WP_001063020 |
| Name | WP_001063020_pUnk_2670 |
UniProt ID | A0A6Y3HD60 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14442.54 Da Isoelectric Point: 5.3125
MAKIQVYVNDHVSEKINAIAVQRRAEGAKEKDVSYSSIASMLLELGLRVYEAQMERKESAFNQAEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGKPKFEYANMVEDIREKVSVEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A6Y3HD60 |
T4CP
| ID | 651 | GenBank | WP_021518367 |
| Name | traD_GKM64_RS20800_pUnk_2670 |
UniProt ID | _ |
| Length | 717 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 717 a.a. Molecular weight: 81500.98 Da Isoelectric Point: 5.0541
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDDKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
KRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQIASLFEPDVPEVVSGEDVTQAEQPQQPVSPAI
NDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPDTQQQMQRREEVN
INVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 652 | GenBank | WP_021518375 |
| Name | traC_GKM64_RS20890_pUnk_2670 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99196.80 Da Isoelectric Point: 5.8486
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSSSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 69415..95718
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GKM64_RS20800 (GKM64_20780) | 69415..71568 | - | 2154 | WP_021518367 | type IV conjugative transfer system coupling protein TraD | virb4 |
| GKM64_RS20805 (GKM64_20785) | 71820..72551 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| GKM64_RS20810 (GKM64_20790) | 72565..73074 | - | 510 | WP_021518368 | conjugal transfer entry exclusion protein TraS | - |
| GKM64_RS20815 (GKM64_20795) | 73071..75896 | - | 2826 | WP_021518369 | conjugal transfer mating-pair stabilization protein TraG | traG |
| GKM64_RS20820 (GKM64_20800) | 75893..77266 | - | 1374 | WP_021518370 | conjugal transfer pilus assembly protein TraH | traH |
| GKM64_RS20825 (GKM64_20805) | 77253..77645 | - | 393 | WP_021518371 | F-type conjugal transfer protein TrbF | - |
| GKM64_RS20830 (GKM64_20810) | 77599..77913 | - | 315 | WP_021518372 | P-type conjugative transfer protein TrbJ | - |
| GKM64_RS20835 (GKM64_20815) | 77903..78457 | - | 555 | WP_001553830 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| GKM64_RS20840 (GKM64_20820) | 78444..78728 | - | 285 | WP_000624105 | type-F conjugative transfer system pilin chaperone TraQ | - |
| GKM64_RS20845 (GKM64_20825) | 78809..79144 | + | 336 | WP_000415571 | hypothetical protein | - |
| GKM64_RS20850 (GKM64_20830) | 79125..79466 | - | 342 | WP_000556794 | conjugal transfer protein TrbA | - |
| GKM64_RS20855 (GKM64_20835) | 79480..80223 | - | 744 | WP_001030378 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| GKM64_RS20860 (GKM64_20840) | 80216..80473 | - | 258 | WP_000864318 | conjugal transfer protein TrbE | - |
| GKM64_RS20865 (GKM64_20845) | 80500..82308 | - | 1809 | WP_021518373 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| GKM64_RS20870 (GKM64_20850) | 82305..82943 | - | 639 | WP_021518374 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| GKM64_RS20875 (GKM64_20855) | 82952..83944 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| GKM64_RS20880 (GKM64_20860) | 83941..84573 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| GKM64_RS20885 (GKM64_20865) | 84570..84956 | - | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| GKM64_RS20890 (GKM64_20870) | 84953..87580 | - | 2628 | WP_021518375 | type IV secretion system protein TraC | virb4 |
| GKM64_RS20895 (GKM64_20875) | 87740..87962 | - | 223 | Protein_85 | conjugal transfer protein TraR | - |
| GKM64_RS20900 (GKM64_20880) | 88097..88612 | - | 516 | WP_000809838 | type IV conjugative transfer system lipoprotein TraV | traV |
| GKM64_RS20905 (GKM64_20885) | 88609..88860 | - | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| GKM64_RS25020 | 88853..89173 | - | 321 | Protein_88 | conjugal transfer protein TrbD | - |
| GKM64_RS20920 (GKM64_20900) | 89160..89750 | - | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| GKM64_RS20925 (GKM64_20905) | 89740..91167 | - | 1428 | WP_015387387 | F-type conjugal transfer pilus assembly protein TraB | traB |
| GKM64_RS20930 (GKM64_20910) | 91167..91895 | - | 729 | WP_001230813 | type-F conjugative transfer system secretin TraK | traK |
| GKM64_RS20935 (GKM64_20915) | 91882..92448 | - | 567 | WP_000399790 | type IV conjugative transfer system protein TraE | traE |
| GKM64_RS20940 (GKM64_20920) | 92470..92781 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| GKM64_RS20945 (GKM64_20925) | 92796..93161 | - | 366 | WP_000340282 | type IV conjugative transfer system pilin TraA | - |
| GKM64_RS20950 (GKM64_20930) | 93204..93419 | - | 216 | WP_001352842 | conjugal transfer relaxosome protein TraY | - |
| GKM64_RS20955 (GKM64_20935) | 93555..94202 | - | 648 | WP_015387377 | conjugal transfer transcriptional regulator TraJ | - |
| GKM64_RS20960 (GKM64_20940) | 94393..94776 | - | 384 | WP_001063020 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| GKM64_RS20965 (GKM64_20945) | 95116..95718 | + | 603 | WP_000243701 | transglycosylase SLT domain-containing protein | - |
| GKM64_RS20970 (GKM64_20950) | 96014..96835 | - | 822 | WP_001513505 | DUF932 domain-containing protein | - |
| GKM64_RS20975 (GKM64_20955) | 96954..97241 | - | 288 | WP_001513506 | hypothetical protein | - |
| GKM64_RS24620 | 97266..97472 | - | 207 | WP_000547955 | hypothetical protein | - |
| GKM64_RS20980 (GKM64_20960) | 97554..97973 | + | 420 | WP_309143924 | hypothetical protein | - |
| GKM64_RS20985 (GKM64_20965) | 98059..98622 | - | 564 | WP_001513507 | class I SAM-dependent methyltransferase | - |
| GKM64_RS20990 (GKM64_20970) | 98670..100031 | - | 1362 | WP_001513508 | DUF3560 domain-containing protein | - |
| GKM64_RS20995 (GKM64_20975) | 100083..100313 | - | 231 | WP_000218642 | hypothetical protein | - |
Host bacterium
| ID | 1501 | GenBank | NZ_WLVN01000005 |
| Plasmid name | pUnk_2670 | Incompatibility group | IncFIB |
| Plasmid size | 162864 bp | Coordinate of oriT [Strand] | 95065..95150 [+] |
| Host baterium | Escherichia coli strain PH-2670-18 |
Cargo genes
| Drug resistance gene | dfrA14, sul2, aph(3'')-Ib, aph(6)-Id, blaTEM-1B, sitABCD |
| Virulence gene | iroB, iroC, iroD, iroE, iroN, iutA, iucC, iucB, iucA |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |