Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100996 |
| Name | oriT_pWI1-incI1 |
| Organism | Escherichia coli isolate WI1 isolate |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_LT838199 (36109..36218 [+], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GTAATTGTAATAGCGTC..GACGGTATTACAATTAC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pWI1-incI1
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file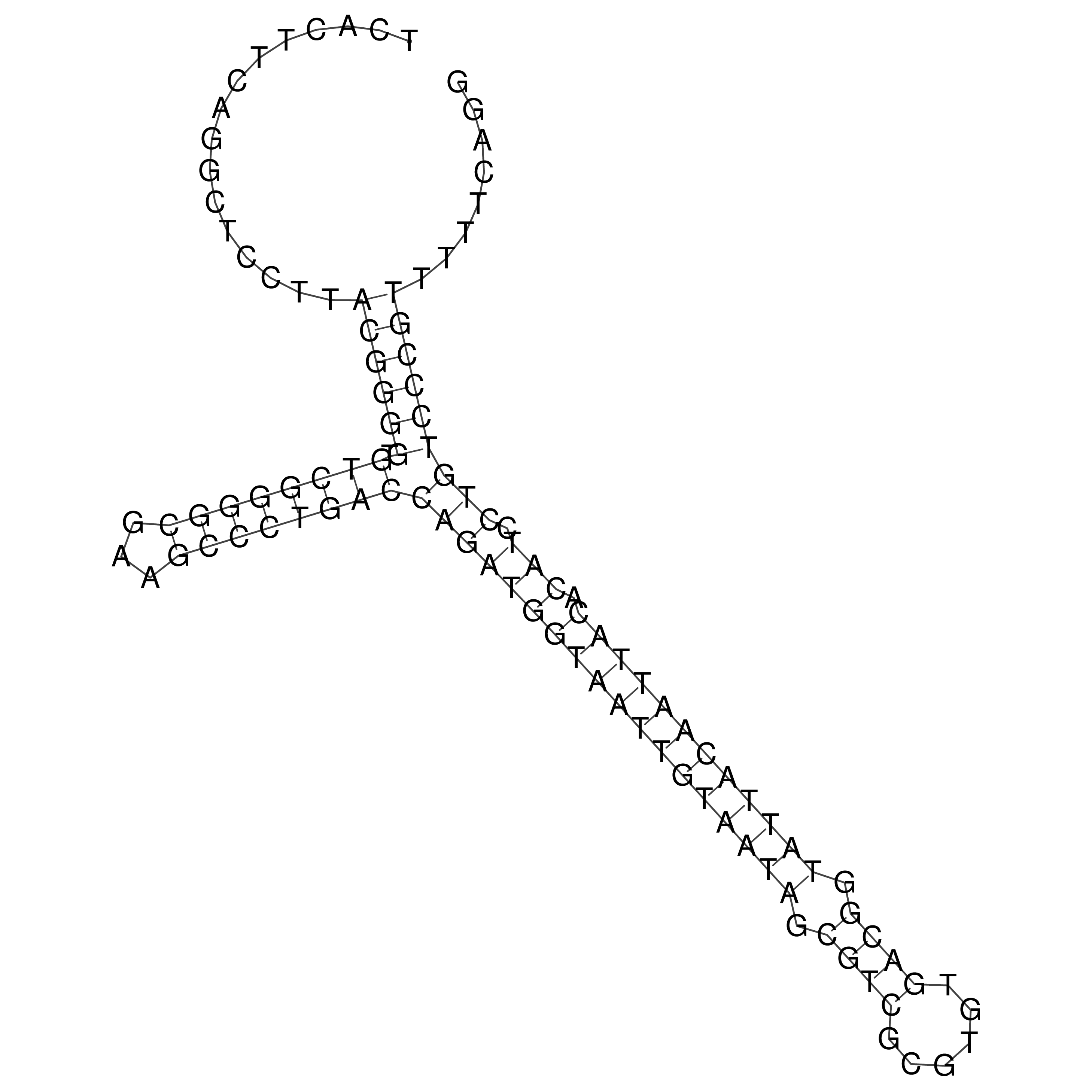
Relaxase
| ID | 1066 | GenBank | WP_001405892 |
| Name | NikB_pWI1-incI1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103883.32 Da Isoelectric Point: 7.3373
>WP_001405892.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1370..30749
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| WI2_RS25495 (ECWI1_P4888) | 65..1219 | + | 1155 | WP_001139957 | site-specific integrase | - |
| WI2_RS25500 (ECWI1_P4889) | 1370..2194 | + | 825 | WP_001238939 | conjugal transfer protein TraE | traE |
| WI2_RS25505 (ECWI1_P4890) | 2279..3481 | + | 1203 | WP_000976351 | conjugal transfer protein TraF | - |
| WI2_RS25510 (ECWI1_P4891) | 3541..4125 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| WI2_RS25515 (ECWI1_P4892) | 4520..4978 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| WI2_RS25520 (ECWI1_P4893) | 4975..5793 | + | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| WI2_RS25525 (ECWI1_P4894) | 5790..6938 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| WI2_RS25530 | 6935..7225 | + | 291 | WP_001299214 | hypothetical protein | traK |
| WI2_RS25535 | 7240..7791 | + | 552 | WP_000014583 | phospholipase D family protein | - |
| WI2_RS25540 (ECWI1_P4896) | 7881..11648 | + | 3768 | WP_044707317 | LPD7 domain-containing protein | - |
| WI2_RS25545 (ECWI1_P4897) | 11666..12013 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| WI2_RS25550 (ECWI1_P4898) | 12010..12702 | + | 693 | WP_000138548 | DotI/IcmL family type IV secretion protein | traM |
| WI2_RS25555 (ECWI1_P4899) | 12713..13696 | + | 984 | WP_001191877 | IncI1-type conjugal transfer protein TraN | traN |
| WI2_RS25560 (ECWI1_P4900) | 13699..14988 | + | 1290 | WP_001272005 | conjugal transfer protein TraO | traO |
| WI2_RS25565 (ECWI1_P4901) | 14988..15692 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| WI2_RS25570 (ECWI1_P4902) | 15692..16219 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| WI2_RS25575 (ECWI1_P4903) | 16270..16674 | + | 405 | WP_000086957 | IncI1-type conjugal transfer protein TraR | traR |
| WI2_RS25580 (ECWI1_P4904) | 16738..16926 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| WI2_RS25585 (ECWI1_P4905) | 16910..17710 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| WI2_RS25590 (ECWI1_P4906) | 17800..20844 | + | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| WI2_RS25595 (ECWI1_P4907) | 20844..21458 | + | 615 | WP_000337399 | IncI1-type conjugal transfer protein TraV | traV |
| WI2_RS25600 (ECWI1_P4908) | 21425..22627 | + | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| WI2_RS25605 (ECWI1_P4909) | 22656..23240 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| WI2_RS25610 (ECWI1_P4910) | 23337..25499 | + | 2163 | WP_000698351 | DotA/TraY family protein | traY |
| WI2_RS25615 (ECWI1_P4911) | 25564..26226 | + | 663 | WP_000653333 | plasmid IncI1-type surface exclusion protein ExcA | - |
| WI2_RS25620 (ECWI1_P4912) | 26298..26507 | - | 210 | WP_000062602 | HEAT repeat domain-containing protein | - |
| WI2_RS26820 | 26850..27026 | + | 177 | WP_001054898 | hypothetical protein | - |
| WI2_RS27415 | 27091..27186 | - | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| WI2_RS25625 (ECWI1_P4915) | 27685..27936 | + | 252 | WP_001291967 | hypothetical protein | - |
| WI2_RS25630 (ECWI1_P4916) | 28008..28160 | - | 153 | WP_001303307 | Hok/Gef family protein | - |
| WI2_RS25635 (ECWI1_P4917) | 28452..29660 | + | 1209 | WP_001303305 | IncI1-type conjugal transfer protein TrbA | trbA |
| WI2_RS25640 (ECWI1_P4918) | 29679..30749 | + | 1071 | WP_085543303 | IncI1-type conjugal transfer protein TrbB | trbB |
| WI2_RS25645 (ECWI1_P4919) | 30742..33033 | + | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 1454 | GenBank | NZ_LT838199 |
| Plasmid name | pWI1-incI1 | Incompatibility group | IncI1 |
| Plasmid size | 83831 bp | Coordinate of oriT [Strand] | 36109..36218 [+] |
| Host baterium | Escherichia coli isolate WI1 isolate |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | Salmonella plasmid virulence |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |