Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100988 |
| Name | oriT_pNDM-JVAP01 |
| Organism | Acinetobacter lactucae strain JVAP01 |
| Sequence Completeness | incomplete |
| NCBI accession of oriT (coordinates [strand]) | NZ_KM923969 (259..296 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 8..17, 21..30 (CATAAGGGAA..TTCCCTTATG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 38 nt
>oriT_pNDM-JVAP01
AGGGATTCATAAGGGAATTATTCCCTTATGTGGGGCTT
AGGGATTCATAAGGGAATTATTCCCTTATGTGGGGCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file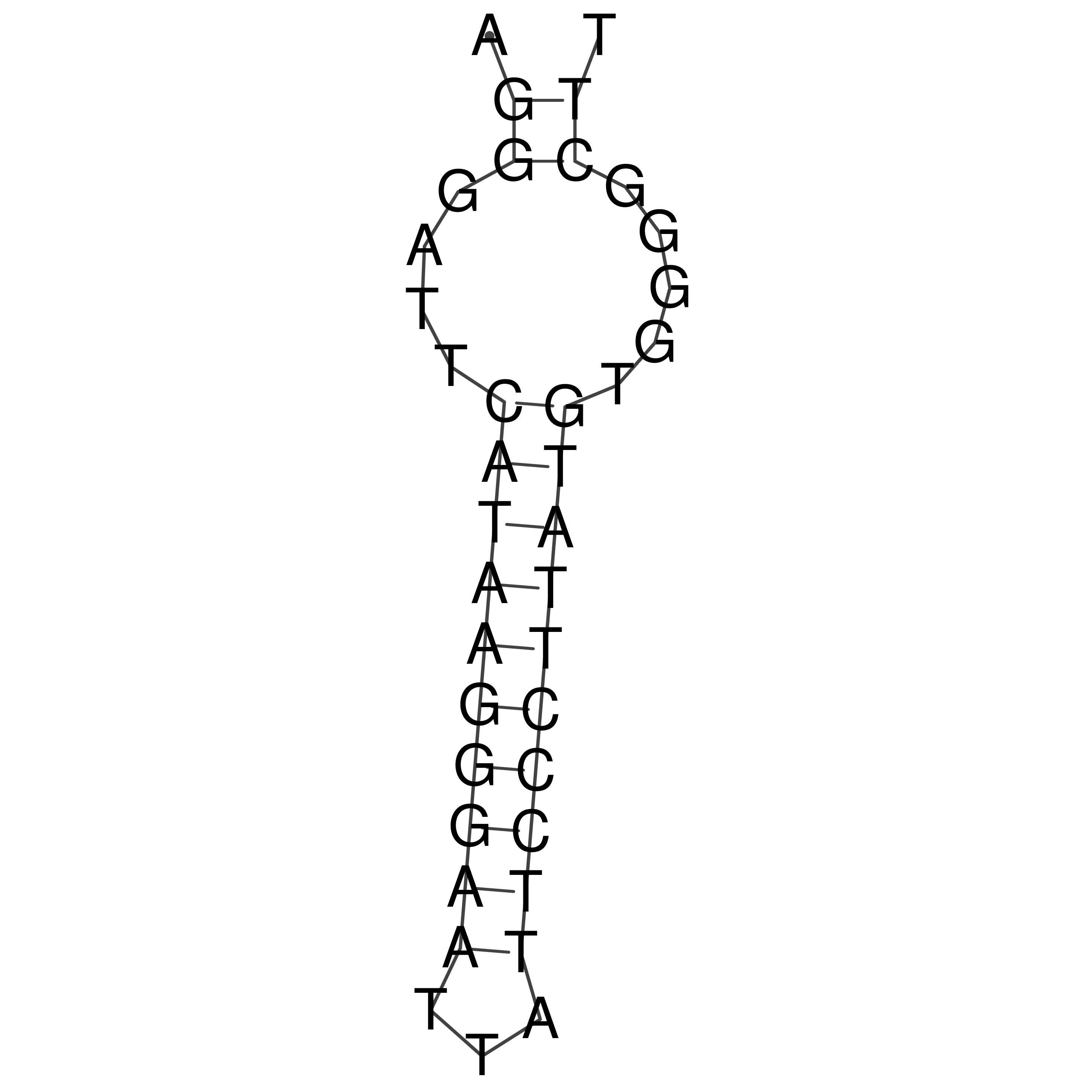
Relaxase
| ID | 1058 | GenBank | WP_017480453 |
| Name | TraA_pNDM-JVAP01 |
UniProt ID | _ |
| Length | 970 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 970 a.a. Molecular weight: 110764.85 Da Isoelectric Point: 8.1822
>WP_017480453.1 MULTISPECIES: AAA family ATPase [Gammaproteobacteria]
MAIYHLSVKTGRQYGSTGTRGANHYAYINRDGKYKRDDLENIKSGNLPSWATDAKHFWKSADKNERINGR
VYTELEISLPRELNKEQREELVEKFVEKTLGENFTYSYAIHSPLASDGQQNPHVHLMFSERKLDGIERDE
VKHFKRYNPVNPEKGGAGKDRYFSSKMFVVDTRLEWANHVNDFCENLGIDARIDHRSFKEQGIELSSQNF
RADYVSSHKYHINENIKQVRHQNGEIIIERPSEAIKALTSNQSVFSVRELERFLMAHTDGEEQYLKAYES
VITCPDMATLYGKDKVVFSSKELVGIEESISTFIYNANEESRIQKPFNLVEKLPLNAVRVAETRTFNSEQ
EKAFYTLTSTDRITLLNGSAGTGKSYVLSAVSEAYKESDYQVYGIALQAITAKAIANDCDIPSSTIASFL
SKYDSGNLEINNKTVLILDEAGMVGSRDMQRLLMLVEKHDAQIKLVGDSYQLSAVSAGHVFANIQENLDK
KNIASLEDIQRQKHAKESDKMIQASKYLSKHEVGKAIDIYKSINMVNEYESHDLALAKTVKSWSEDSSES
KLMLAHSNSDVNELNRMARALLIKQGKLNPESVSVKNYQGDLDISIGEKIVFKQNNSQMKVSNGELAKVI
GFEKDKFNNPKAMMVQMESDSRIVRVDLKEYDQFKHGYANTIHSSQGMTVDSAYIVASENMNANLTYVAL
TRHKQNIQINYSALTFATNEKETVKTPDGKVSYDHNYKPVEREVTAFENFKKVLGRSETKEFSTDFTLVE
SKDNLMKSYLNDHRLHLENKRELFGINSKLYSLTSENKTFTKEEIVSEVRASLKSKALLGDEIVKFNGNM
NIAKGELVKLTNDLEIKTGLFKKVTFEKGTELKVIDAYKVKDKPVMKARIKNEEYEIPFNKLNLEYSDKY
FNEFKDQSKARFDSRHKTEVYTEFREQLLKQKANAPKDNLNAPNQRIKNPNTPNRNTPKY
MAIYHLSVKTGRQYGSTGTRGANHYAYINRDGKYKRDDLENIKSGNLPSWATDAKHFWKSADKNERINGR
VYTELEISLPRELNKEQREELVEKFVEKTLGENFTYSYAIHSPLASDGQQNPHVHLMFSERKLDGIERDE
VKHFKRYNPVNPEKGGAGKDRYFSSKMFVVDTRLEWANHVNDFCENLGIDARIDHRSFKEQGIELSSQNF
RADYVSSHKYHINENIKQVRHQNGEIIIERPSEAIKALTSNQSVFSVRELERFLMAHTDGEEQYLKAYES
VITCPDMATLYGKDKVVFSSKELVGIEESISTFIYNANEESRIQKPFNLVEKLPLNAVRVAETRTFNSEQ
EKAFYTLTSTDRITLLNGSAGTGKSYVLSAVSEAYKESDYQVYGIALQAITAKAIANDCDIPSSTIASFL
SKYDSGNLEINNKTVLILDEAGMVGSRDMQRLLMLVEKHDAQIKLVGDSYQLSAVSAGHVFANIQENLDK
KNIASLEDIQRQKHAKESDKMIQASKYLSKHEVGKAIDIYKSINMVNEYESHDLALAKTVKSWSEDSSES
KLMLAHSNSDVNELNRMARALLIKQGKLNPESVSVKNYQGDLDISIGEKIVFKQNNSQMKVSNGELAKVI
GFEKDKFNNPKAMMVQMESDSRIVRVDLKEYDQFKHGYANTIHSSQGMTVDSAYIVASENMNANLTYVAL
TRHKQNIQINYSALTFATNEKETVKTPDGKVSYDHNYKPVEREVTAFENFKKVLGRSETKEFSTDFTLVE
SKDNLMKSYLNDHRLHLENKRELFGINSKLYSLTSENKTFTKEEIVSEVRASLKSKALLGDEIVKFNGNM
NIAKGELVKLTNDLEIKTGLFKKVTFEKGTELKVIDAYKVKDKPVMKARIKNEEYEIPFNKLNLEYSDKY
FNEFKDQSKARFDSRHKTEVYTEFREQLLKQKANAPKDNLNAPNQRIKNPNTPNRNTPKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22716..36011
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A3Q30_RS00100 (JVAP01_p24) | 18594..19241 | - | 648 | WP_004993315 | recombinase family protein | - |
| A3Q30_RS00105 | 19393..19617 | + | 225 | WP_015056389 | hypothetical protein | - |
| A3Q30_RS18365 (JVAP01_p26) | 19801..21513 | + | 1713 | WP_015060705 | zeta toxin family protein | - |
| A3Q30_RS00115 (JVAP01_p27) | 21510..21896 | - | 387 | WP_227513993 | thermonuclease family protein | - |
| A3Q30_RS00120 (JVAP01_p28) | 22192..22653 | - | 462 | WP_015060707 | hypothetical protein | - |
| A3Q30_RS00125 (JVAP01_p29) | 22716..23357 | - | 642 | WP_004999358 | type IV secretion system protein | virB5 |
| A3Q30_RS00130 (JVAP01_p30) | 23354..26065 | - | 2712 | WP_015060708 | VirB4 | virb4 |
| A3Q30_RS00135 (JVAP01_p31) | 26077..26370 | - | 294 | WP_017480461 | TrbC/VirB2 family protein | virB2 |
| A3Q30_RS00140 (JVAP01_p32) | 26381..27091 | - | 711 | WP_017480462 | lytic transglycosylase domain-containing protein | - |
| A3Q30_RS00145 (JVAP01_p33) | 27136..27336 | - | 201 | WP_004999346 | hypothetical protein | - |
| A3Q30_RS00150 (JVAP01_p34) | 27355..28872 | - | 1518 | WP_015060710 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| A3Q30_RS00155 (JVAP01_p35) | 28869..30116 | - | 1248 | WP_017480463 | lytic transglycosylase domain-containing protein | - |
| A3Q30_RS00160 (JVAP01_p36) | 30198..30650 | - | 453 | WP_004999340 | hypothetical protein | - |
| A3Q30_RS00165 (JVAP01_p37) | 30654..30983 | - | 330 | WP_004999338 | hypothetical protein | - |
| A3Q30_RS00170 (JVAP01_p38) | 30952..31989 | - | 1038 | WP_015060713 | P-type DNA transfer ATPase VirB11 | virB11 |
| A3Q30_RS00175 (JVAP01_p39) | 32005..33384 | - | 1380 | WP_005000433 | TrbI/VirB10 family protein | virB10 |
| A3Q30_RS00180 (JVAP01_p40) | 33386..34123 | - | 738 | WP_005000435 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| A3Q30_RS00185 (JVAP01_p41) | 34138..34926 | - | 789 | WP_005000436 | VirB8/TrbF family protein | virB8 |
| A3Q30_RS00190 (JVAP01_p42) | 34926..36011 | - | 1086 | WP_005000437 | type IV secretion system protein | virB6 |
| A3Q30_RS00195 (JVAP01_p43) | 36019..36438 | - | 420 | WP_005000439 | hypothetical protein | - |
| A3Q30_RS00200 (JVAP01_p44) | 36425..36796 | - | 372 | WP_005000442 | hypothetical protein | - |
| A3Q30_RS00210 (JVAP01_p46) | 38418..38657 | + | 240 | WP_005000443 | hypothetical protein | - |
| A3Q30_RS00215 (JVAP01_p47) | 38674..39363 | + | 690 | WP_005000446 | ParA family partition ATPase | - |
| A3Q30_RS00220 (JVAP01_p48) | 39360..39764 | + | 405 | WP_005000449 | hypothetical protein | - |
Host bacterium
| ID | 1446 | GenBank | NZ_KM923969 |
| Plasmid name | pNDM-JVAP01 | Incompatibility group | _ |
| Plasmid size | 47268 bp | Coordinate of oriT [Strand] | 259..296 [+] |
| Host baterium | Acinetobacter lactucae strain JVAP01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |