Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100965 |
| Name | oriT_tig00000001_p1 |
| Organism | Klebsiella pneumoniae strain AR_0143 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021709 (1..458 [+], 458 nt) |
| oriT length | 458 nt |
| IRs (inverted repeats) | 50..57, 61..68 (TGGCCTGC..GGAGGCCA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 458 nt
>oriT_tig00000001_p1
AAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAAGGCTAATCAGGAGCCTGTGATTTACCCGATCATTGAAGAAGCTCTGCATCGCTACAGCCAGCTCGTGTTTCATGAGCAGAGAGAGAAATATGAAGACCCGGCCAGAATTGGGGCATTTCTGGAAACCCTGATCACCGAAACCTGCCGGGCGTTGGAAGTGCAAATTGTCGATAGTGGCGGTGATTCATGGTCTGTCGATTCAGGAGAGTCGTTCTCACTGTGGCTTTCTTCCCATCCAGGAGAACTATCCATTAACCCGCAGCCCCATGAGGATGAGACCTCTTTGCGTGGCTTGCTGTATGAGCTCATCACCTGTGAGAGCGTGAAAACTGTTTTAAGGAGAACCGACT
AAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAAGGCTAATCAGGAGCCTGTGATTTACCCGATCATTGAAGAAGCTCTGCATCGCTACAGCCAGCTCGTGTTTCATGAGCAGAGAGAGAAATATGAAGACCCGGCCAGAATTGGGGCATTTCTGGAAACCCTGATCACCGAAACCTGCCGGGCGTTGGAAGTGCAAATTGTCGATAGTGGCGGTGATTCATGGTCTGTCGATTCAGGAGAGTCGTTCTCACTGTGGCTTTCTTCCCATCCAGGAGAACTATCCATTAACCCGCAGCCCCATGAGGATGAGACCTCTTTGCGTGGCTTGCTGTATGAGCTCATCACCTGTGAGAGCGTGAAAACTGTTTTAAGGAGAACCGACT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file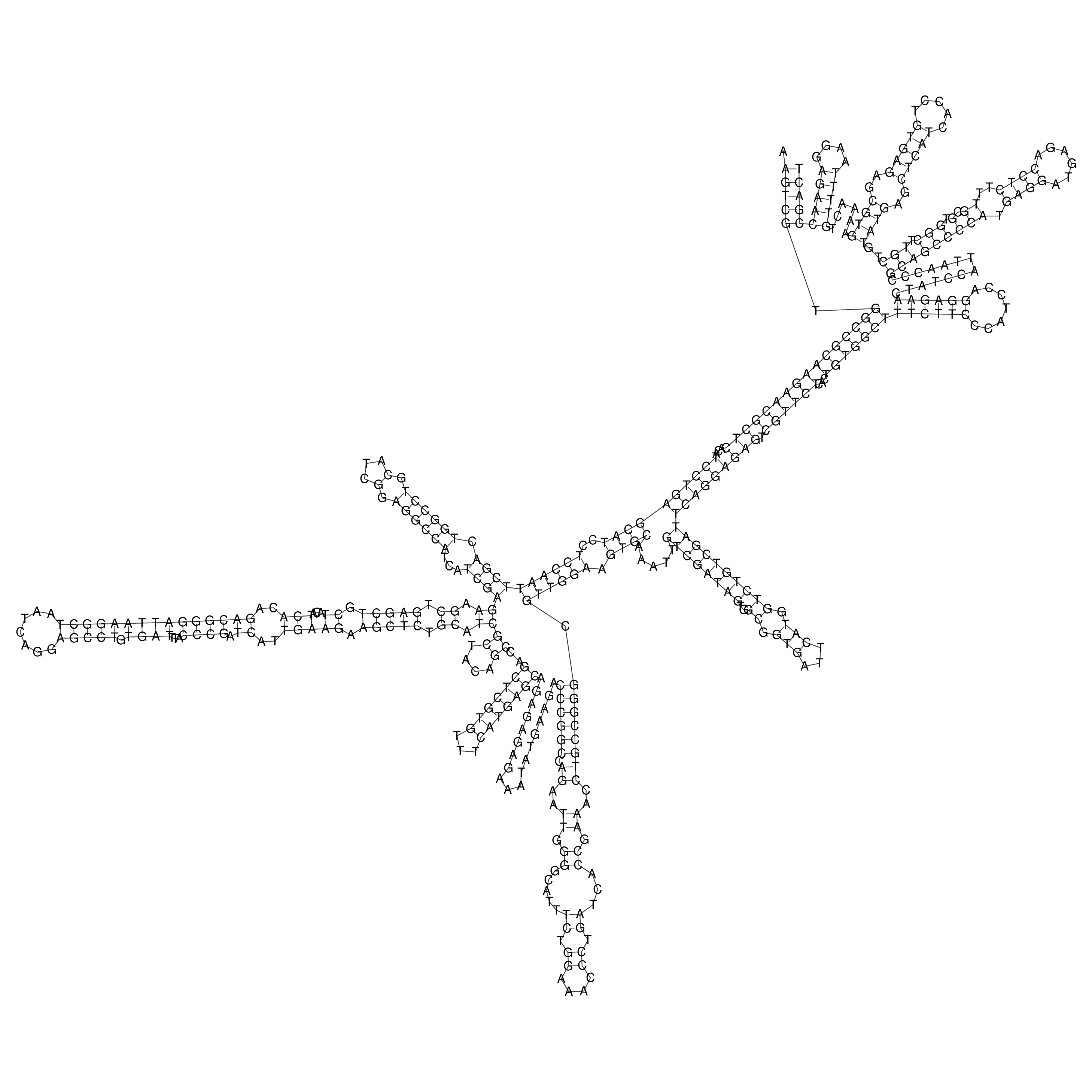
Relaxase
| ID | 1035 | GenBank | WP_001326173 |
| Name | TraI_tig00000001_p1 |
UniProt ID | _ |
| Length | 990 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 990 a.a. Molecular weight: 109356.90 Da Isoelectric Point: 4.9395
>WP_001326173.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALNKLFGGRSGVIETAPSVRVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEVDDDQEEDVSALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGSTESSKPD
AGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDA
FSASAETASTDATNSEIPDVAMPGKQEKQPKQDFVPQEQNSLQGDDFPMFGSSDEPPSWAIEPLPMLTDA
PEQTTPAPAMPPTDKPNLHEKDAKTLLVEMLAGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVILY
PDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQDA
FELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKPSPEQKAKGKDSQPQQKEKKVDVTSPVEEPQRQ
PVQEKQNVARLPKREVQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSGVT
PQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKKRQ
GKLYLEVNET
MLKALNKLFGGRSGVIETAPSVRVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEVDDDQEEDVSALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGSTESSKPD
AGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPFDA
FSASAETASTDATNSEIPDVAMPGKQEKQPKQDFVPQEQNSLQGDDFPMFGSSDEPPSWAIEPLPMLTDA
PEQTTPAPAMPPTDKPNLHEKDAKTLLVEMLAGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVILY
PDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQDA
FELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKPSPEQKAKGKDSQPQQKEKKVDVTSPVEEPQRQ
PVQEKQNVARLPKREVQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSGVT
PQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKKRQ
GKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 459..7132
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AM390_RS27085 (AM390_26950) | 459..1088 | + | 630 | WP_000743449 | DUF4400 domain-containing protein | tfc7 |
| AM390_RS27090 (AM390_26955) | 1098..1544 | + | 447 | WP_000122507 | hypothetical protein | - |
| AM390_RS27095 (AM390_26960) | 1554..1931 | + | 378 | WP_000869297 | hypothetical protein | - |
| AM390_RS27100 (AM390_26965) | 1931..2593 | + | 663 | WP_001231464 | hypothetical protein | - |
| AM390_RS31280 | 2774..2917 | + | 144 | WP_001275801 | hypothetical protein | - |
| AM390_RS27105 (AM390_26970) | 2929..3294 | + | 366 | WP_001052530 | hypothetical protein | - |
| AM390_RS27110 (AM390_26975) | 3439..3720 | + | 282 | WP_000805625 | type IV conjugative transfer system protein TraL | traL |
| AM390_RS27115 (AM390_26980) | 3717..4343 | + | 627 | WP_001049717 | TraE/TraK family type IV conjugative transfer system protein | traE |
| AM390_RS27120 (AM390_26985) | 4327..5244 | + | 918 | WP_000794249 | type-F conjugative transfer system secretin TraK | traK |
| AM390_RS27125 (AM390_26990) | 5244..6557 | + | 1314 | WP_024131605 | TraB/VirB10 family protein | traB |
| AM390_RS27130 (AM390_26995) | 6554..7132 | + | 579 | WP_000793435 | type IV conjugative transfer system lipoprotein TraV | traV |
| AM390_RS27135 (AM390_27000) | 7136..7528 | + | 393 | WP_000479535 | TraA family conjugative transfer protein | - |
| AM390_RS27140 (AM390_27005) | 7813..8985 | + | 1173 | Protein_13 | IS1380-like element ISEcp1 family transposase | - |
| AM390_RS27145 (AM390_27010) | 9091..10278 | + | 1188 | WP_080399009 | IS4 family transposase | - |
| AM390_RS27150 (AM390_27015) | 10733..11878 | + | 1146 | WP_015056382 | class C beta-lactamase CMY-4 | - |
Host bacterium
| ID | 1423 | GenBank | NZ_CP021709 |
| Plasmid name | tig00000001_p1 | Incompatibility group | IncA/C2 |
| Plasmid size | 186758 bp | Coordinate of oriT [Strand] | 1..458 [+] |
| Host baterium | Klebsiella pneumoniae strain AR_0143 |
Cargo genes
| Drug resistance gene | resistance to cetylpyridinium; quaternary ammonium compound resistance |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |