Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100928 |
| Name | oriT_pCFSAN004174 |
| Organism | Salmonella enterica subsp. enterica serovar Saintpaul strain CFSAN004174 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019207 (27278..27387 [+], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pCFSAN004174
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file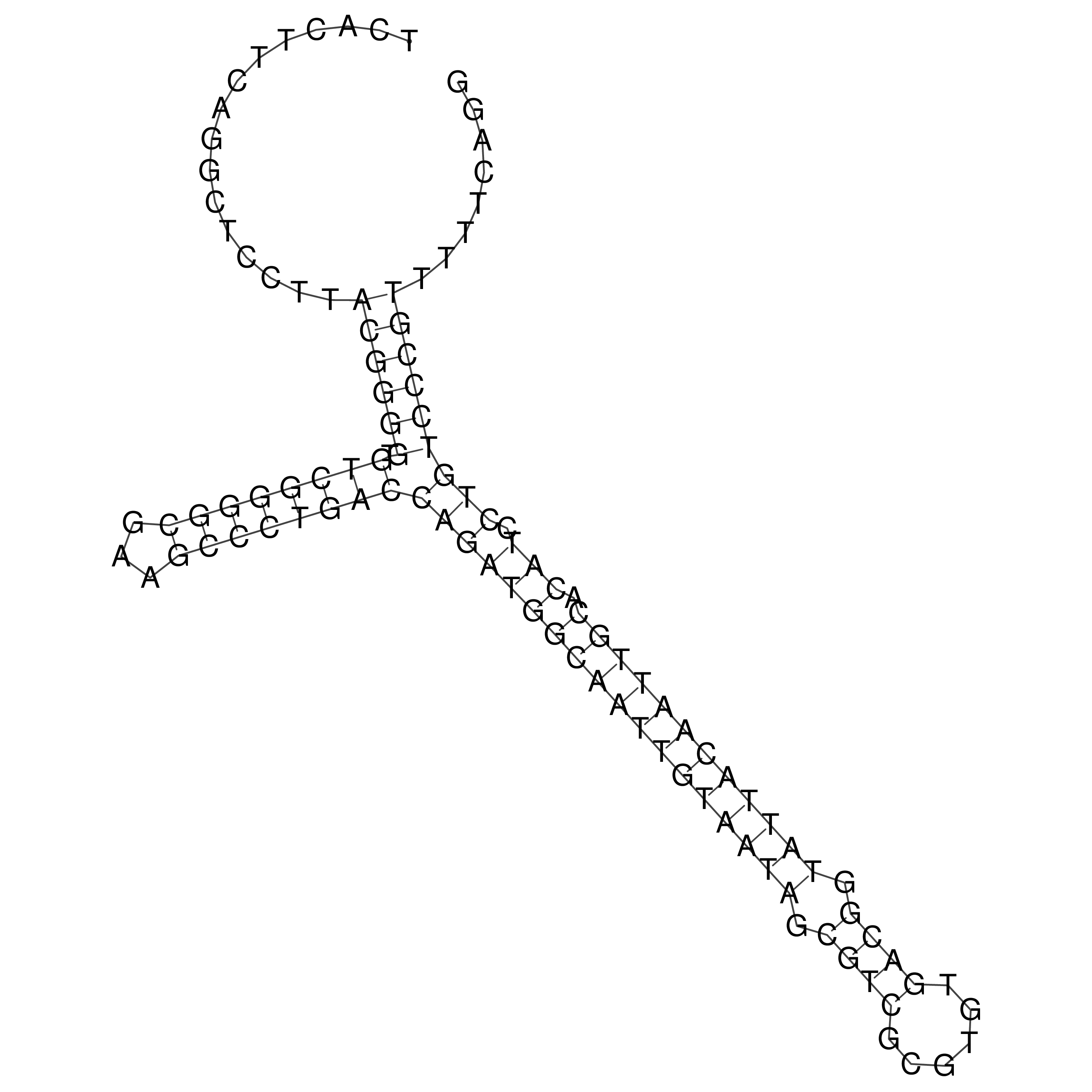
Relaxase
| ID | 999 | GenBank | WP_076033867 |
| Name | NikB_pCFSAN004174 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104039.45 Da Isoelectric Point: 7.3426
>WP_076033867.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDVFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDVFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2782..21917
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BKQ23_RS23965 (BKQ23_23965) | 2782..3129 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| BKQ23_RS23970 (BKQ23_23970) | 3126..3818 | + | 693 | WP_000138551 | DotI/IcmL family type IV secretion protein | traM |
| BKQ23_RS23975 (BKQ23_23975) | 3829..4812 | + | 984 | WP_001191880 | IncI1-type conjugal transfer protein TraN | traN |
| BKQ23_RS23980 (BKQ23_23980) | 4815..6104 | + | 1290 | WP_023196135 | conjugal transfer protein TraO | traO |
| BKQ23_RS23985 (BKQ23_23985) | 6104..6808 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| BKQ23_RS23990 (BKQ23_23990) | 6808..7335 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| BKQ23_RS23995 (BKQ23_23995) | 7386..7790 | + | 405 | WP_001422824 | IncI1-type conjugal transfer protein TraR | traR |
| BKQ23_RS24000 (BKQ23_24000) | 7854..8042 | + | 189 | WP_001277253 | putative conjugal transfer protein TraS | - |
| BKQ23_RS24005 (BKQ23_24005) | 8026..8826 | + | 801 | WP_001164783 | IncI1-type conjugal transfer protein TraT | traT |
| BKQ23_RS24010 (BKQ23_24010) | 8916..11960 | + | 3045 | WP_021265678 | IncI1-type conjugal transfer protein TraU | traU |
| BKQ23_RS24015 (BKQ23_24015) | 11960..12574 | + | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| BKQ23_RS24020 (BKQ23_24020) | 12541..13743 | + | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| BKQ23_RS24025 (BKQ23_24025) | 13772..14356 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| BKQ23_RS24030 (BKQ23_24030) | 14453..16621 | + | 2169 | WP_000698357 | DotA/TraY family protein | traY |
| BKQ23_RS24035 (BKQ23_24035) | 16695..17345 | + | 651 | WP_001178506 | plasmid IncI1-type surface exclusion protein ExcA | - |
| BKQ23_RS24040 (BKQ23_24040) | 17417..17626 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| BKQ23_RS25240 | 18018..18194 | + | 177 | WP_001054904 | hypothetical protein | - |
| BKQ23_RS25560 | 18259..18354 | - | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| BKQ23_RS24045 (BKQ23_24045) | 18853..19104 | + | 252 | WP_001291964 | hypothetical protein | - |
| BKQ23_RS24050 (BKQ23_24050) | 19176..19328 | - | 153 | WP_001303307 | Hok/Gef family protein | - |
| BKQ23_RS24055 (BKQ23_24055) | 19620..20828 | + | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| BKQ23_RS24060 (BKQ23_24060) | 20847..21917 | + | 1071 | WP_001749135 | IncI1-type conjugal transfer protein TrbB | trbB |
| BKQ23_RS25460 (BKQ23_24065) | 21910..24202 | + | 2293 | Protein_23 | F-type conjugative transfer protein TrbC | - |
Region 2: 67552..79754
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BKQ23_RS24385 (BKQ23_24385) | 62614..63681 | + | 1068 | WP_001545759 | type IV pilus biogenesis lipoprotein PilL | - |
| BKQ23_RS24390 (BKQ23_24390) | 63681..64118 | + | 438 | WP_001545758 | type IV pilus biogenesis protein PilM | - |
| BKQ23_RS24395 (BKQ23_24395) | 64132..65814 | + | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| BKQ23_RS24400 (BKQ23_24400) | 65807..67102 | + | 1296 | WP_016245572 | type 4b pilus protein PilO2 | - |
| BKQ23_RS24405 (BKQ23_24405) | 67089..67541 | + | 453 | WP_001247333 | type IV pilus biogenesis protein PilP | - |
| BKQ23_RS24410 (BKQ23_24410) | 67552..69105 | + | 1554 | WP_001545756 | ATPase, T2SS/T4P/T4SS family | virB11 |
| BKQ23_RS24415 (BKQ23_24415) | 69118..70203 | + | 1086 | WP_001208802 | type II secretion system F family protein | - |
| BKQ23_RS24420 (BKQ23_24420) | 70220..70834 | + | 615 | WP_000908227 | type 4 pilus major pilin | - |
| BKQ23_RS24425 (BKQ23_24425) | 70844..71404 | + | 561 | WP_000014005 | lytic transglycosylase domain-containing protein | virB1 |
| BKQ23_RS24430 (BKQ23_24430) | 71389..72045 | + | 657 | WP_001193549 | A24 family peptidase | - |
| BKQ23_RS24435 (BKQ23_24435) | 72045..73337 | + | 1293 | WP_024145563 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| BKQ23_RS24440 (BKQ23_24440) | 73394..73651 | - | 258 | Protein_84 | transposase | - |
| BKQ23_RS25615 | 73655..73747 | + | 93 | Protein_85 | shufflon-specific DNA recombinase | - |
| BKQ23_RS24450 (BKQ23_24450) | 73898..74722 | + | 825 | WP_076033864 | conjugal transfer protein TraE | traE |
| BKQ23_RS24455 (BKQ23_24455) | 74808..76010 | + | 1203 | WP_023196138 | conjugal transfer protein TraF | - |
| BKQ23_RS24460 (BKQ23_24460) | 76070..76654 | + | 585 | WP_001611722 | histidine phosphatase family protein | - |
| BKQ23_RS24465 (BKQ23_24465) | 77049..77507 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| BKQ23_RS24470 (BKQ23_24470) | 77504..78322 | + | 819 | WP_076033865 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| BKQ23_RS24475 (BKQ23_24475) | 78319..79467 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| BKQ23_RS24480 (BKQ23_24480) | 79464..79754 | + | 291 | WP_001314267 | hypothetical protein | traK |
| BKQ23_RS24485 (BKQ23_24485) | 79769..80320 | + | 552 | WP_000014586 | phospholipase D family protein | - |
Host bacterium
| ID | 1387 | GenBank | NZ_CP019207 |
| Plasmid name | pCFSAN004174 | Incompatibility group | IncI1 |
| Plasmid size | 81410 bp | Coordinate of oriT [Strand] | 27278..27387 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Saintpaul strain CFSAN004174 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |