Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100923 |
| Name | oriT_pEC244_2 |
| Organism | Escherichia coli strain Ecol_244 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019018 (105284..105573 [+], 290 nt) |
| oriT length | 290 nt |
| IRs (inverted repeats) | 225..232, 235..242 n (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 251..252 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 290 nt
>oriT_pEC244_2
CGCACCGCTAGCAGCGCCCCTAGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCTCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAACTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
CGCACCGCTAGCAGCGCCCCTAGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCTCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAACTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file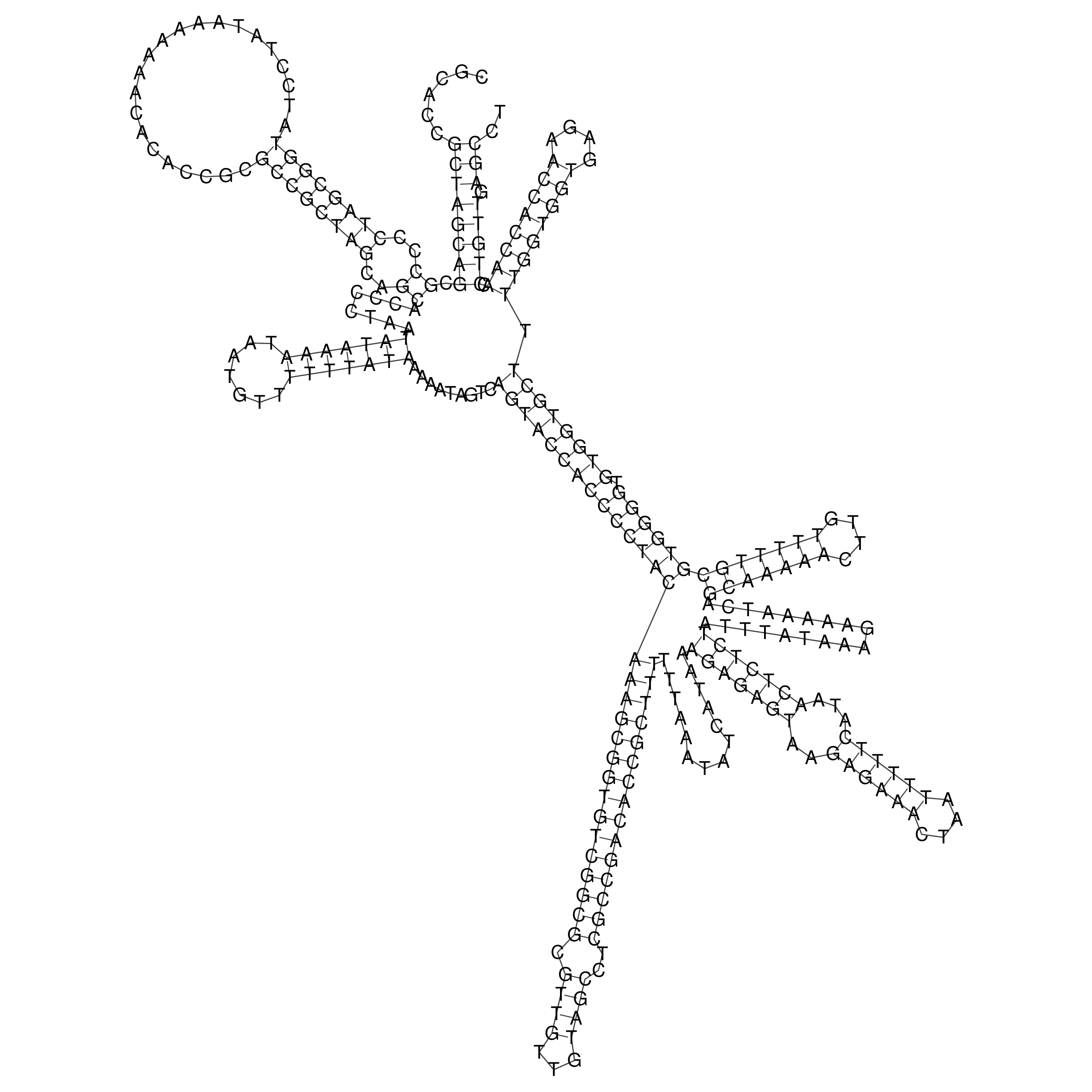
Relaxase
| ID | 994 | GenBank | WP_001611029 |
| Name | TraI_pEC244_2 |
UniProt ID | A0A075MCD1 |
| Length | 1756 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191983.83 Da Isoelectric Point: 5.9594
>WP_001611029.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGLPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFIDLGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRECGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMTGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVVAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPRTITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVRDIAGLERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGLPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFIDLGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRECGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMTGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVVAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPRTITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVRDIAGLERDRSAISEREAALPESVLREPQRVREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 79690..106135
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BE937_RS01160 (BE937_01230) | 79056..79283 | + | 228 | WP_000450520 | toxin-antitoxin system antitoxin VapB | - |
| BE937_RS01165 (BE937_01235) | 79283..79681 | + | 399 | WP_000911313 | type II toxin-antitoxin system VapC family toxin | - |
| BE937_RS01170 (BE937_01240) | 79690..81915 | - | 2226 | WP_064237182 | type IV conjugative transfer system coupling protein TraD | virb4 |
| BE937_RS01175 (BE937_01245) | 82168..82899 | - | 732 | WP_023565183 | conjugal transfer complement resistance protein TraT | - |
| BE937_RS01180 (BE937_01250) | 82931..83428 | - | 498 | WP_000605857 | entry exclusion protein | - |
| BE937_RS01185 (BE937_01255) | 83444..86266 | - | 2823 | WP_001007018 | conjugal transfer mating-pair stabilization protein TraG | traG |
| BE937_RS01190 (BE937_01260) | 86263..87636 | - | 1374 | WP_001517790 | conjugal transfer pilus assembly protein TraH | traH |
| BE937_RS01195 (BE937_01265) | 87636..88016 | - | 381 | WP_001680290 | F-type conjugal transfer protein TrbF | - |
| BE937_RS01200 (BE937_01270) | 88003..88344 | - | 342 | WP_001443031 | P-type conjugative transfer protein TrbJ | - |
| BE937_RS01205 (BE937_01275) | 88274..88819 | - | 546 | WP_001680291 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| BE937_RS01210 (BE937_01280) | 88806..89090 | - | 285 | WP_000624109 | type-F conjugative transfer system pilin chaperone TraQ | - |
| BE937_RS01215 (BE937_01285) | 89171..89485 | + | 315 | WP_000415566 | protein ArtA | - |
| BE937_RS01220 (BE937_01290) | 89487..89834 | - | 348 | WP_001287910 | conjugal transfer protein TrbA | - |
| BE937_RS01225 (BE937_01295) | 89850..90593 | - | 744 | WP_001680292 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| BE937_RS01230 (BE937_01300) | 90586..90843 | - | 258 | WP_000864318 | conjugal transfer protein TrbE | - |
| BE937_RS01235 (BE937_01305) | 90870..92678 | - | 1809 | WP_000821849 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| BE937_RS01240 (BE937_01310) | 92675..93313 | - | 639 | WP_000777695 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| BE937_RS01245 (BE937_01315) | 93322..94314 | - | 993 | WP_000830180 | conjugal transfer pilus assembly protein TraU | traU |
| BE937_RS01250 (BE937_01320) | 94311..94943 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| BE937_RS01255 (BE937_01325) | 94940..95326 | - | 387 | WP_001680293 | type-F conjugative transfer system protein TrbI | - |
| BE937_RS01260 (BE937_01330) | 95323..97950 | - | 2628 | WP_001064245 | type IV secretion system protein TraC | virb4 |
| BE937_RS01265 (BE937_01335) | 98110..98331 | - | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| BE937_RS01270 (BE937_01340) | 98466..98981 | - | 516 | WP_000809838 | type IV conjugative transfer system lipoprotein TraV | traV |
| BE937_RS01275 (BE937_01345) | 98978..99229 | - | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| BE937_RS01280 (BE937_01350) | 99241..99438 | - | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| BE937_RS01285 (BE937_01355) | 99425..100015 | - | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| BE937_RS01290 (BE937_01360) | 100005..101432 | - | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| BE937_RS01295 (BE937_01365) | 101432..102160 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| BE937_RS01300 (BE937_01370) | 102147..102713 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| BE937_RS01305 (BE937_01375) | 102735..103046 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| BE937_RS01310 (BE937_01380) | 103061..103426 | - | 366 | WP_000994779 | type IV conjugative transfer system pilin TraA | - |
| BE937_RS01315 (BE937_01385) | 103459..103854 | - | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| BE937_RS01320 (BE937_01390) | 103953..104642 | - | 690 | WP_000283385 | conjugal transfer transcriptional regulator TraJ | - |
| BE937_RS01325 (BE937_01395) | 104829..105212 | - | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| BE937_RS01330 (BE937_01400) | 105545..106135 | + | 591 | WP_279947734 | transglycosylase SLT domain-containing protein | virB1 |
| BE937_RS01335 (BE937_01405) | 106432..107253 | - | 822 | WP_001234487 | DUF932 domain-containing protein | - |
| BE937_RS01340 (BE937_01410) | 107364..107660 | - | 297 | WP_001272245 | hypothetical protein | - |
| BE937_RS28830 | 107712..107985 | + | 274 | Protein_112 | hypothetical protein | - |
| BE937_RS01350 (BE937_01420) | 108286..108411 | - | 126 | WP_096937776 | type I toxin-antitoxin system Hok family toxin | - |
| BE937_RS28835 | 108353..108502 | - | 150 | Protein_114 | plasmid maintenance protein Mok | - |
| BE937_RS01355 (BE937_01430) | 108681..109443 | - | 763 | Protein_115 | plasmid SOS inhibition protein A | - |
| BE937_RS01360 (BE937_01435) | 109440..109874 | - | 435 | WP_000845962 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 1382 | GenBank | NZ_CP019018 |
| Plasmid name | pEC244_2 | Incompatibility group | IncFIB |
| Plasmid size | 158911 bp | Coordinate of oriT [Strand] | 105284..105573 [+] |
| Host baterium | Escherichia coli strain Ecol_244 |
Cargo genes
| Drug resistance gene | sitABCD |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |