Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100914 |
| Name | oriT_pEC545_1 |
| Organism | Escherichia coli strain Ecol_545 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP018975 (39695..39804 [+], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pEC545_1
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file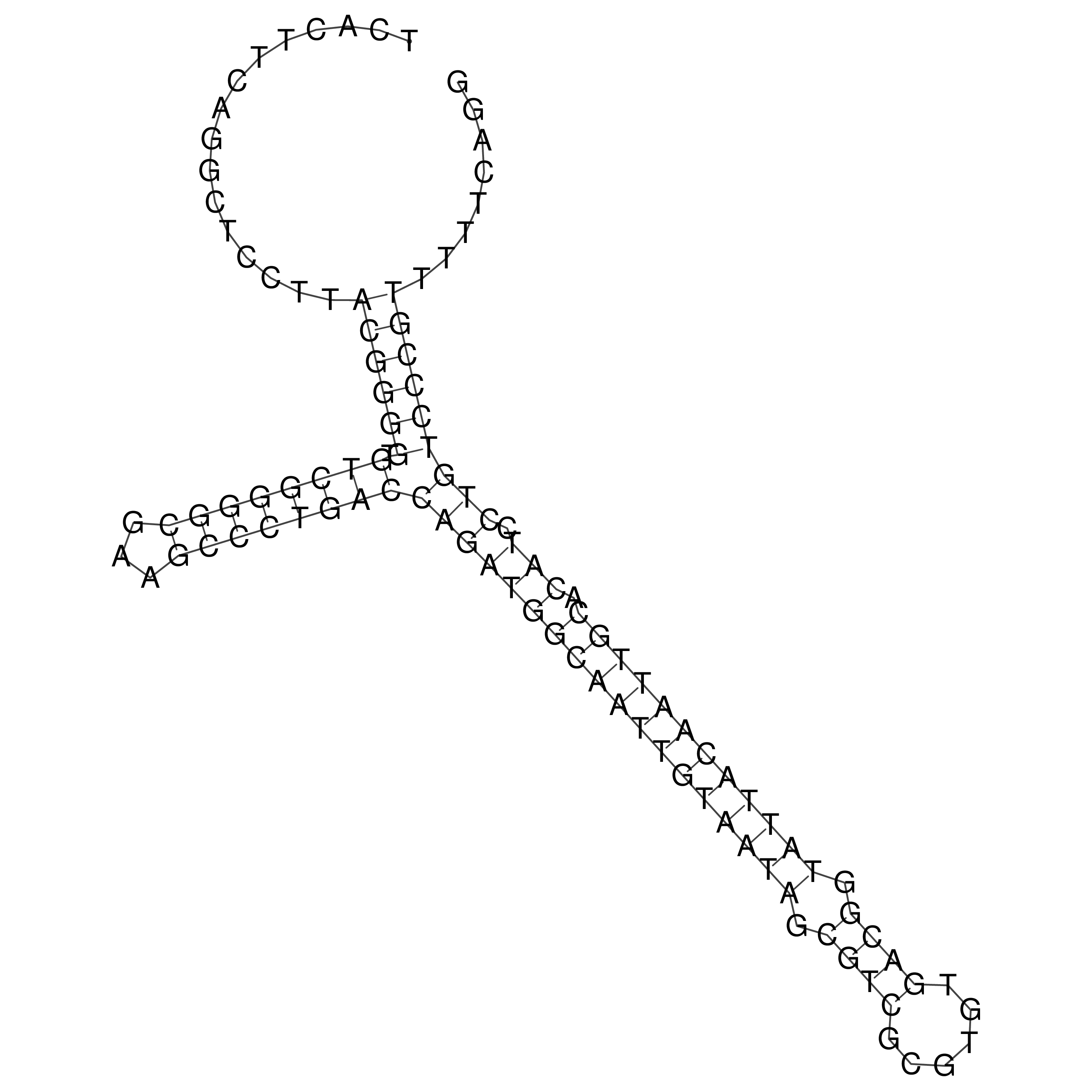
Relaxase
| ID | 985 | GenBank | WP_012783930 |
| Name | Relaxase_pEC545_1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104070.55 Da Isoelectric Point: 7.3623
>WP_012783930.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALVDKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALVDKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3472..34335
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BE949_RS28375 (BE949_01040) | 1031..1255 | - | 225 | Protein_1 | hypothetical protein | - |
| BE949_RS28065 | 1815..2168 | - | 354 | WP_001393368 | hypothetical protein | - |
| BE949_RS01015 (BE949_01065) | 2167..3321 | + | 1155 | WP_001139955 | site-specific integrase | - |
| BE949_RS01020 (BE949_01070) | 3472..4296 | + | 825 | WP_001238932 | conjugal transfer protein TraE | traE |
| BE949_RS01025 (BE949_01075) | 4382..5584 | + | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| BE949_RS01030 (BE949_01080) | 5644..6228 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| BE949_RS01035 (BE949_01085) | 6622..7080 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| BE949_RS01040 (BE949_01090) | 7077..7895 | + | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| BE949_RS01045 (BE949_01095) | 7892..9040 | + | 1149 | WP_001024971 | plasmid transfer ATPase TraJ | virB11 |
| BE949_RS01050 (BE949_01100) | 9037..9327 | + | 291 | WP_001314267 | hypothetical protein | traK |
| BE949_RS01055 (BE949_01105) | 9342..9893 | + | 552 | WP_012783942 | phospholipase D family protein | - |
| BE949_RS01060 (BE949_01110) | 9983..13750 | + | 3768 | WP_012783940 | LPD7 domain-containing protein | - |
| BE949_RS01065 (BE949_01115) | 13768..14115 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| BE949_RS01070 (BE949_01120) | 14112..14804 | + | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| BE949_RS01075 (BE949_01125) | 14815..15798 | + | 984 | WP_001191880 | IncI1-type conjugal transfer protein TraN | traN |
| BE949_RS01080 (BE949_01130) | 15801..17090 | + | 1290 | WP_011264042 | conjugal transfer protein TraO | traO |
| BE949_RS01085 (BE949_01135) | 17090..17794 | + | 705 | WP_001493811 | IncI1-type conjugal transfer protein TraP | traP |
| BE949_RS01090 (BE949_01140) | 17794..18321 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| BE949_RS01095 (BE949_01145) | 18372..18776 | + | 405 | WP_000086965 | IncI1-type conjugal transfer protein TraR | traR |
| BE949_RS01100 (BE949_01150) | 18840..19028 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| BE949_RS01105 (BE949_01155) | 19012..19812 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| BE949_RS01110 (BE949_01160) | 19902..22946 | + | 3045 | WP_012783939 | IncI1-type conjugal transfer protein TraU | traU |
| BE949_RS01115 (BE949_01165) | 22946..23560 | + | 615 | WP_000337394 | IncI1-type conjugal transfer protein TraV | traV |
| BE949_RS01120 (BE949_01170) | 23527..24729 | + | 1203 | WP_012783938 | IncI1-type conjugal transfer protein TraW | traW |
| BE949_RS01125 (BE949_01175) | 24758..25342 | + | 585 | WP_012783937 | conjugal transfer protein TraX | - |
| BE949_RS01130 (BE949_01180) | 25439..27607 | + | 2169 | WP_015508354 | DotA/TraY family protein | traY |
| BE949_RS01135 (BE949_01185) | 27678..28340 | + | 663 | WP_012783935 | plasmid IncI1-type surface exclusion protein ExcA | - |
| BE949_RS01140 (BE949_01190) | 28412..28621 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| BE949_RS27745 (BE949_01200) | 29013..29189 | + | 177 | WP_001054900 | hypothetical protein | - |
| BE949_RS28305 | 29254..29349 | - | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| BE949_RS01150 (BE949_01210) | 29850..30101 | + | 252 | WP_001291964 | hypothetical protein | - |
| BE949_RS01155 (BE949_01215) | 30173..30325 | - | 153 | WP_001387489 | Hok/Gef family protein | - |
| BE949_RS01160 (BE949_01220) | 30952..31731 | + | 780 | WP_275450201 | protein FinQ | - |
| BE949_RS01165 (BE949_01225) | 32038..33246 | + | 1209 | WP_001703845 | IncI1-type conjugal transfer protein TrbA | trbA |
| BE949_RS01170 (BE949_01230) | 33265..34335 | + | 1071 | WP_012783932 | IncI1-type conjugal transfer protein TrbB | trbB |
| BE949_RS01175 (BE949_01235) | 34328..36619 | + | 2292 | WP_012783931 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 1373 | GenBank | NZ_CP018975 |
| Plasmid name | pEC545_1 | Incompatibility group | IncI1 |
| Plasmid size | 95926 bp | Coordinate of oriT [Strand] | 39695..39804 [+] |
| Host baterium | Escherichia coli strain Ecol_545 |
Cargo genes
| Drug resistance gene | blaTEM-1B, blaCTX-M-15 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrVA2 |