Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100869 |
| Name | oriT_pMR0716_IncFII |
| Organism | Escherichia coli strain MRSN352231 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP018107 (24436..24905 [-], 470 nt) |
| oriT length | 470 nt |
| IRs (inverted repeats) | 142..151, 154..163 (ATAGTGTCGT..ACGACCCTAT) 231..238, 241..248 (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 257..258 |
| Conserved sequence flanking the nic site |
GTAGTGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 470 nt
>oriT_pMR0716_IncFII
AAGAAGAGCACCCCTAGCAGCGCCCCTAGGGTCATACTATAAAAAAACGCACGGCGCTGCTAGGGGCGGCCCTAATATATCCAAATGTTTTTCATAAAATTTGTCAGTACTGACCCTAACAAGGGTCGCCATAGGGTCGCCATAGGGTCGTCAACGACCCTATTTAATAATGTAAAATAAATTAAAATACATTATTAAAAACATAAGCTAATGATTCAAAAAGCAAATCAGCAAAAACTTGTTTTTGCGTAGTGTGTGGTGCTTTTGGTGGTGAGAACCACCAACTAGTTGAGCCTTATAGTGGAGTAGGTTAAATTATTTCCGGATAATGTCACCAGAGGTGAAAAAATGAAAAAGTGGATGTTAGCAATCTTCCTGATGTTTATAAGTGGGATCTGTGAAGCCGCCGATTGTTTTGATCTTGCAGGCCGGGATTACAAAATAGACCCGGATTTACTGAGAGCGATATC
AAGAAGAGCACCCCTAGCAGCGCCCCTAGGGTCATACTATAAAAAAACGCACGGCGCTGCTAGGGGCGGCCCTAATATATCCAAATGTTTTTCATAAAATTTGTCAGTACTGACCCTAACAAGGGTCGCCATAGGGTCGCCATAGGGTCGTCAACGACCCTATTTAATAATGTAAAATAAATTAAAATACATTATTAAAAACATAAGCTAATGATTCAAAAAGCAAATCAGCAAAAACTTGTTTTTGCGTAGTGTGTGGTGCTTTTGGTGGTGAGAACCACCAACTAGTTGAGCCTTATAGTGGAGTAGGTTAAATTATTTCCGGATAATGTCACCAGAGGTGAAAAAATGAAAAAGTGGATGTTAGCAATCTTCCTGATGTTTATAAGTGGGATCTGTGAAGCCGCCGATTGTTTTGATCTTGCAGGCCGGGATTACAAAATAGACCCGGATTTACTGAGAGCGATATC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file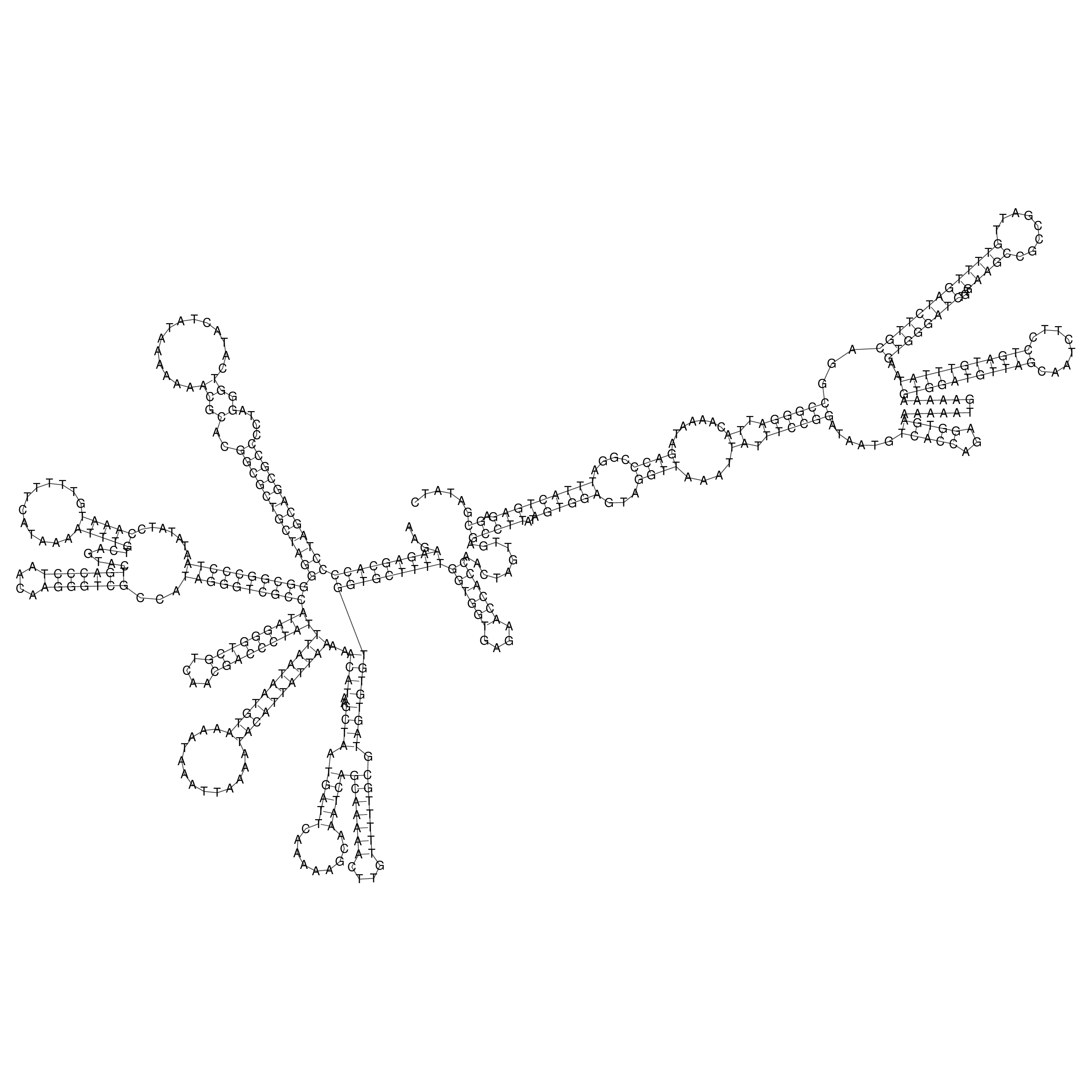
Relaxase
| ID | 940 | GenBank | WP_071940847 |
| Name | TraI_pMR0716_IncFII |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191890.47 Da Isoelectric Point: 6.0275
>WP_071940847.1 conjugative transfer relaxase/helicase TraI [Escherichia coli]
MLSFSVVKSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFSENVLANRIAFGKIYQSELRQRVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRGAQGETQVVRISSLDS
SWSLFRPEKMPVAGGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPATLPVSNSPFTALKL
ESGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFVDLGREINA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYTLIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQEAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVS
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPAQEQAEQHIDLAYAI
TAHGAQGASETFAIVLEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAGQERDRAAITEREAALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
MLSFSVVKSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFSENVLANRIAFGKIYQSELRQRVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRGAQGETQVVRISSLDS
SWSLFRPEKMPVAGGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPATLPVSNSPFTALKL
ESGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFVDLGREINA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYTLIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQEAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVS
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPAQEQAEQHIDLAYAI
TAHGAQGASETFAIVLEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAGQERDRAAITEREAALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 26966..51444
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BSG22_RS30565 (BSG22_25680) | 22297..22503 | + | 207 | WP_000547945 | hypothetical protein | - |
| BSG22_RS27520 (BSG22_25685) | 22527..22823 | + | 297 | WP_001272239 | hypothetical protein | - |
| BSG22_RS27525 (BSG22_25690) | 22929..23750 | + | 822 | WP_001444560 | DUF932 domain-containing protein | - |
| BSG22_RS27530 (BSG22_25695) | 24048..24617 | - | 570 | Protein_42 | transglycosylase SLT domain-containing protein | - |
| BSG22_RS27535 (BSG22_25700) | 24972..25355 | + | 384 | WP_029397922 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| BSG22_RS27540 (BSG22_25705) | 25546..26232 | + | 687 | WP_029397923 | PAS domain-containing protein | - |
| BSG22_RS27545 (BSG22_25710) | 26326..26553 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| BSG22_RS27550 (BSG22_25715) | 26587..26952 | + | 366 | WP_000340274 | type IV conjugative transfer system pilin TraA | - |
| BSG22_RS27555 (BSG22_25720) | 26966..27277 | + | 312 | WP_071940841 | type IV conjugative transfer system protein TraL | traL |
| BSG22_RS27560 (BSG22_25725) | 27299..27865 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| BSG22_RS27565 (BSG22_25730) | 27852..28580 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| BSG22_RS27570 (BSG22_25735) | 28580..30007 | + | 1428 | WP_000146636 | F-type conjugal transfer pilus assembly protein TraB | traB |
| BSG22_RS27575 (BSG22_25740) | 29997..30584 | + | 588 | WP_071940842 | conjugal transfer pilus-stabilizing protein TraP | - |
| BSG22_RS27580 (BSG22_25745) | 30571..30888 | + | 318 | WP_024189706 | conjugal transfer protein TrbD | - |
| BSG22_RS27585 (BSG22_25750) | 30888..31403 | + | 516 | WP_071940843 | type IV conjugative transfer system lipoprotein TraV | traV |
| BSG22_RS27590 | 31538..31759 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| BSG22_RS27595 (BSG22_25755) | 31919..34546 | + | 2628 | WP_071940844 | type IV secretion system protein TraC | virb4 |
| BSG22_RS27600 (BSG22_25760) | 34543..34929 | + | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| BSG22_RS27605 (BSG22_25765) | 34926..35558 | + | 633 | WP_062873969 | type-F conjugative transfer system protein TraW | traW |
| BSG22_RS27610 (BSG22_25770) | 35555..36547 | + | 993 | WP_021559016 | conjugal transfer pilus assembly protein TraU | traU |
| BSG22_RS27615 (BSG22_25775) | 36577..36882 | + | 306 | WP_029397934 | hypothetical protein | - |
| BSG22_RS27620 (BSG22_25780) | 36891..37529 | + | 639 | WP_029397935 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| BSG22_RS27625 (BSG22_25785) | 37526..37897 | + | 372 | WP_000056336 | hypothetical protein | - |
| BSG22_RS27630 (BSG22_25790) | 37923..38345 | + | 423 | WP_001229309 | HNH endonuclease signature motif containing protein | - |
| BSG22_RS27635 (BSG22_25795) | 38342..40192 | + | 1851 | WP_071940845 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| BSG22_RS27640 (BSG22_25800) | 40219..40476 | + | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| BSG22_RS27645 (BSG22_25805) | 40469..41212 | + | 744 | WP_001030371 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| BSG22_RS27650 (BSG22_25810) | 41373..41609 | + | 237 | WP_000270010 | type-F conjugative transfer system pilin chaperone TraQ | - |
| BSG22_RS27655 (BSG22_25815) | 41596..42141 | + | 546 | WP_000059818 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| BSG22_RS27660 | 42131..42445 | + | 315 | WP_001384255 | P-type conjugative transfer protein TrbJ | - |
| BSG22_RS27665 (BSG22_25820) | 42399..42791 | + | 393 | WP_053913402 | F-type conjugal transfer protein TrbF | - |
| BSG22_RS27670 (BSG22_25825) | 42778..44151 | + | 1374 | WP_021532658 | conjugal transfer pilus assembly protein TraH | traH |
| BSG22_RS27675 (BSG22_25830) | 44148..46970 | + | 2823 | WP_069040907 | conjugal transfer mating-pair stabilization protein TraG | traG |
| BSG22_RS27680 (BSG22_25835) | 46986..47483 | + | 498 | WP_000605867 | hypothetical protein | - |
| BSG22_RS27685 (BSG22_25840) | 47515..48246 | + | 732 | WP_021559024 | conjugal transfer complement resistance protein TraT | - |
| BSG22_RS27690 (BSG22_25845) | 48449..49186 | + | 738 | WP_000199911 | hypothetical protein | - |
| BSG22_RS27695 (BSG22_25850) | 49237..51444 | + | 2208 | WP_071940846 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 1328 | GenBank | NZ_CP018107 |
| Plasmid name | pMR0716_IncFII | Incompatibility group | IncFII(29) |
| Plasmid size | 61085 bp | Coordinate of oriT [Strand] | 24436..24905 [-] |
| Host baterium | Escherichia coli strain MRSN352231 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |