Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100864 |
| Name | oriT_pCAV1015-76 |
| Organism | Klebsiella oxytoca strain CAV1015 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017932 (72439..72543 [+], 105 nt) |
| oriT length | 105 nt |
| IRs (inverted repeats) | 18..35, 42..59 (GTTTTTGGTACACCGCCG..CGGCAGTGACGCAAAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 105 nt
>oriT_pCAV1015-76
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file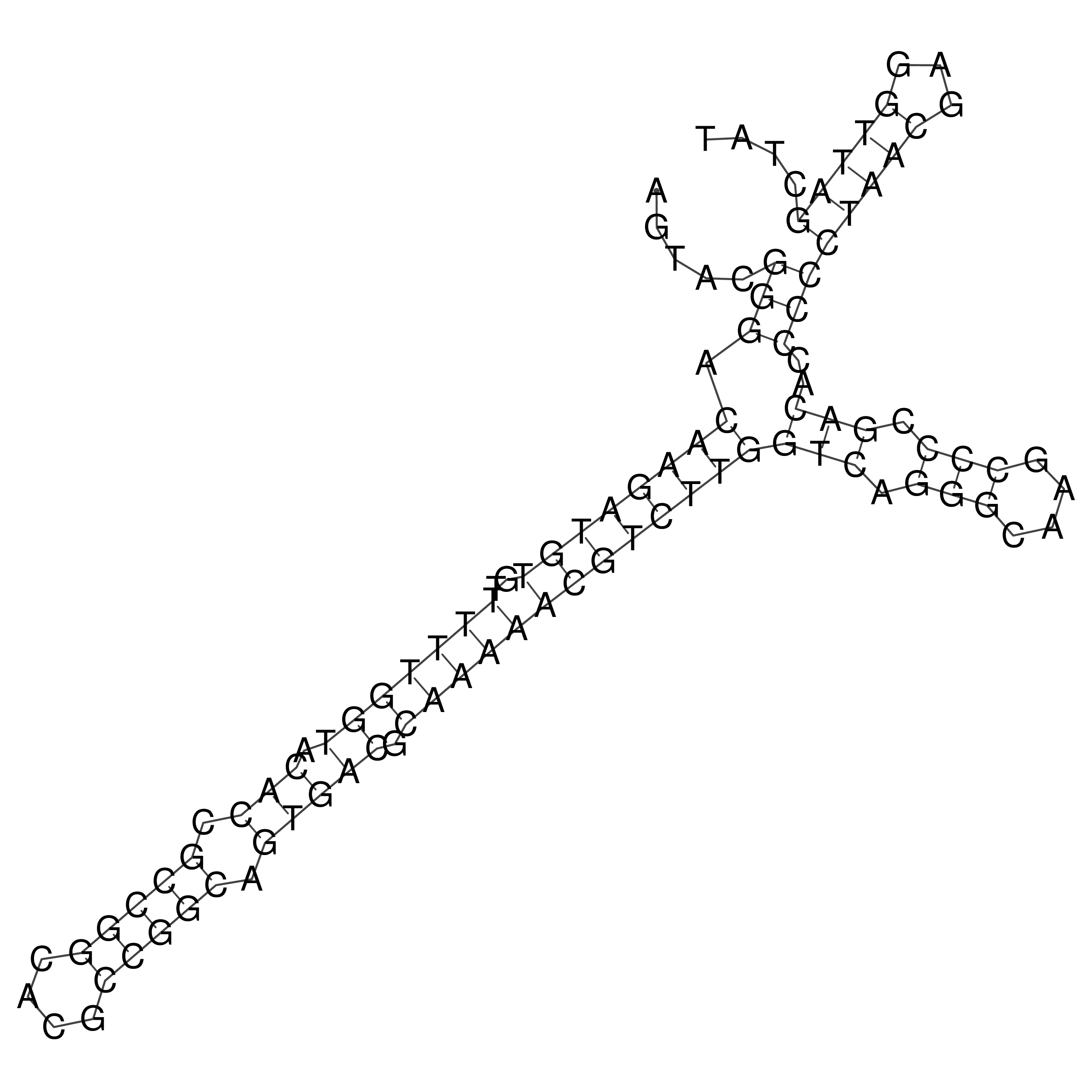
Relaxase
| ID | 936 | GenBank | WP_004206924 |
| Name | PutativerelaxaseofpCAV1015-76 |
UniProt ID | _ |
| Length | 659 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75087.84 Da Isoelectric Point: 10.0285
>WP_004206924.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacterales]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQLRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQLRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1098..18351
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AGF18_RS30610 (AGF18_30610) | 1098..1358 | + | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| AGF18_RS33920 | 1383..4688 | + | 3306 | WP_227516140 | LPD7 domain-containing protein | - |
| AGF18_RS30620 (AGF18_30620) | 4654..5166 | + | 513 | WP_011091071 | hypothetical protein | traL |
| AGF18_RS32555 | 5167..5379 | - | 213 | WP_305953721 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| AGF18_RS30625 (AGF18_30625) | 5351..5761 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| AGF18_RS30630 (AGF18_30630) | 5829..6542 | + | 714 | WP_223294958 | DotI/IcmL family type IV secretion protein | traM |
| AGF18_RS32560 | 6551..7702 | + | 1152 | WP_015062843 | DotH/IcmK family type IV secretion protein | traN |
| AGF18_RS30645 (AGF18_30645) | 7714..9063 | + | 1350 | WP_004206932 | conjugal transfer protein TraO | traO |
| AGF18_RS30650 (AGF18_30650) | 9075..9779 | + | 705 | WP_004206933 | conjugal transfer protein TraP | traP |
| AGF18_RS30655 (AGF18_30655) | 9803..10333 | + | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| AGF18_RS30660 (AGF18_30660) | 10350..10739 | + | 390 | WP_011154472 | DUF6750 family protein | traR |
| AGF18_RS30665 (AGF18_30665) | 10785..11279 | + | 495 | WP_004187480 | hypothetical protein | - |
| AGF18_RS30670 (AGF18_30670) | 11276..14326 | + | 3051 | WP_011154474 | conjugative transfer protein | traU |
| AGF18_RS30675 (AGF18_30675) | 14323..15531 | + | 1209 | WP_004187486 | conjugal transfer protein TraW | traW |
| AGF18_RS30680 (AGF18_30680) | 15528..16178 | + | 651 | WP_004187488 | hypothetical protein | - |
| AGF18_RS30685 (AGF18_30685) | 16171..18351 | + | 2181 | WP_004187492 | DotA/TraY family protein | traY |
| AGF18_RS30690 (AGF18_30690) | 18354..19007 | + | 654 | WP_004206935 | hypothetical protein | - |
| AGF18_RS30695 (AGF18_30695) | 19081..19311 | + | 231 | WP_004187496 | IncL/M type plasmid replication protein RepC | - |
| AGF18_RS30700 (AGF18_30700) | 19608..20663 | + | 1056 | WP_015062846 | plasmid replication initiator RepA | - |
| AGF18_RS30705 (AGF18_30705) | 21835..22368 | - | 534 | Protein_20 | IS110-like element IS4321 family transposase | - |
Host bacterium
| ID | 1324 | GenBank | NZ_CP017932 |
| Plasmid name | pCAV1015-76 | Incompatibility group | IncL/M |
| Plasmid size | 76186 bp | Coordinate of oriT [Strand] | 72439..72543 [+] |
| Host baterium | Klebsiella oxytoca strain CAV1015 |
Cargo genes
| Drug resistance gene | aac(6')-Ib-cr, blaOXA-1, catB3, ARR-3, qacE, sul1, blaSHV-30 |
| Virulence gene | - |
| Metal resistance gene | merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |