Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100856 |
| Name | oriT_pSARB26_02 |
| Organism | Salmonella enterica subsp. enterica serovar Stanleyville str. CFSAN000624 strain SGSC 2518 isolate SARB61 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017725 (15892..16540 [+], 649 nt) |
| oriT length | 649 nt |
| IRs (inverted repeats) | IR1: 198..207, 211..220 (ATTAGGTGTT..AATACCTAAT) IR2:319..324, 330..335 (AAAAAA..TTTTTT) IR3: 393..402, 405..414 (GTATTCATGC..GCATGAATAC) |
| Location of nic site | 301..302 |
| Conserved sequence flanking the nic site |
GGTGT|ATAGC |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 649 nt
>oriT_pSARB26_02
AGCGCCGCAGATAATCTGACCGATTACCTCCTGAAACCAGGTCTATATAGGCCAAAAGTTCATCTGATACTTTTGCGGTTATTATTGGCATTCAGTCCTCACATTGTGCATTTCTTAAACAAAAGATTGGGATCTAACAAGCTGAAATCTTAGTATTACCAAAGTAATAAAGCAAACTCATTATAAAACAATGAGTTATTAGGTGTTTTTAATACCTAATTATTACCGAATATTGACGCTATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAGGAGCGATCTACGTAGGTTAAGGACTAACTGACTAAAAAGCGTTCAATATTCCGTATTCATGCTTGCATGAATACCAGTACAACACTATTACAACAAAAGTACATCAAAATTACATCAAAAGTACATCACTTGAAGGTTGACAGTACAACAGAATTACATCATTATCTGGTCATGAGGTAGCCAGTACAACAAAAGTACATCAAAAATACATCATAAATACATCAGAAATACATCAAAATTACATCATTCTAAATGAGGGTACTATGAAGCCCAAAAGTATCAGGGCGGCACTTCAGTTGATGTTGCCGG
AGCGCCGCAGATAATCTGACCGATTACCTCCTGAAACCAGGTCTATATAGGCCAAAAGTTCATCTGATACTTTTGCGGTTATTATTGGCATTCAGTCCTCACATTGTGCATTTCTTAAACAAAAGATTGGGATCTAACAAGCTGAAATCTTAGTATTACCAAAGTAATAAAGCAAACTCATTATAAAACAATGAGTTATTAGGTGTTTTTAATACCTAATTATTACCGAATATTGACGCTATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAGGAGCGATCTACGTAGGTTAAGGACTAACTGACTAAAAAGCGTTCAATATTCCGTATTCATGCTTGCATGAATACCAGTACAACACTATTACAACAAAAGTACATCAAAATTACATCAAAAGTACATCACTTGAAGGTTGACAGTACAACAGAATTACATCATTATCTGGTCATGAGGTAGCCAGTACAACAAAAGTACATCAAAAATACATCATAAATACATCAGAAATACATCAAAATTACATCATTCTAAATGAGGGTACTATGAAGCCCAAAAGTATCAGGGCGGCACTTCAGTTGATGTTGCCGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file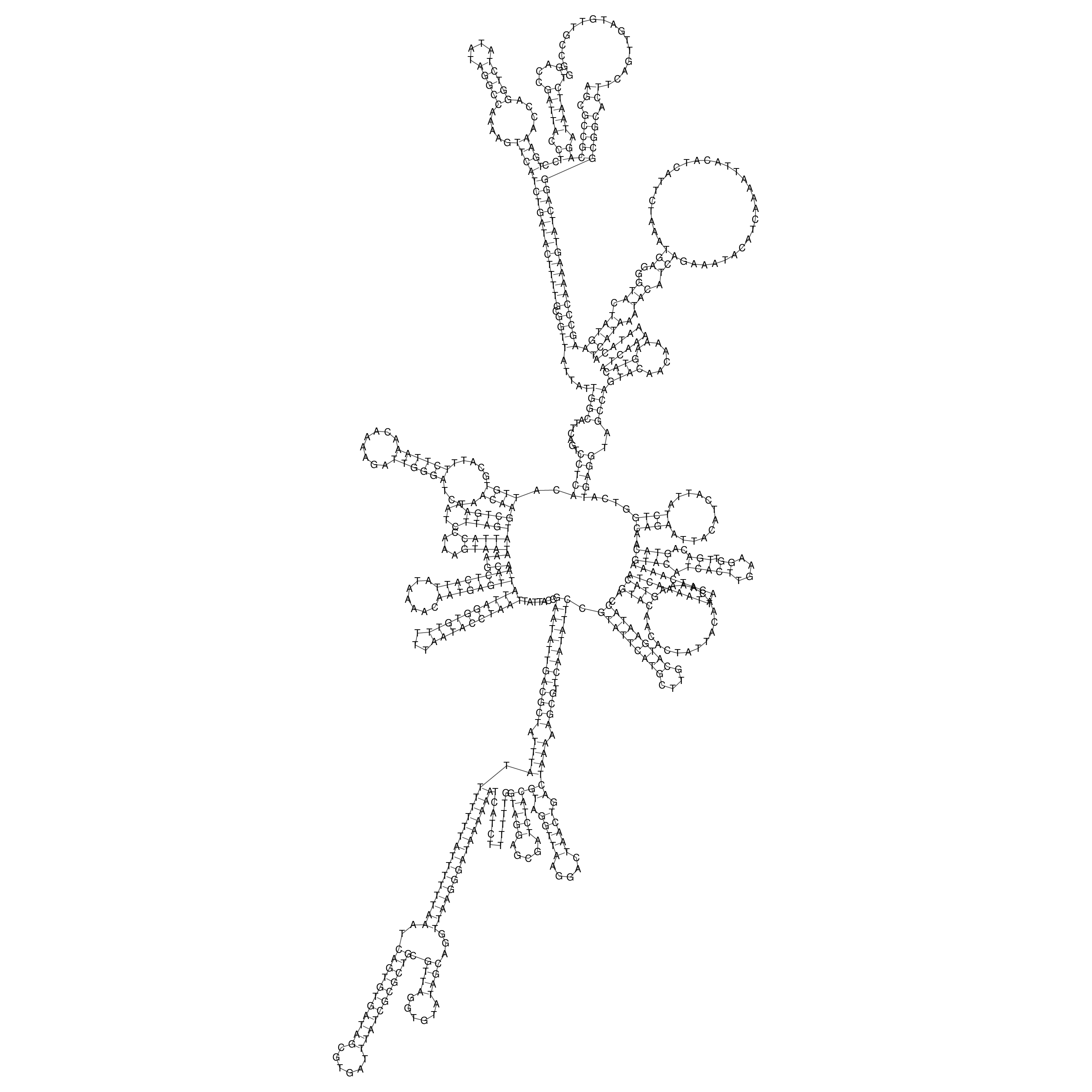
Relaxase
| ID | 928 | GenBank | WP_000884352 |
| Name | Relaxase_pSARB26_02 |
UniProt ID | _ |
| Length | 1078 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120163.41 Da Isoelectric Point: 6.5766
>WP_000884352.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36534..46859
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CFSAN000624_RS24530 (CFSAN000624_024530) | 32385..33818 | - | 1434 | WP_001288435 | DNA cytosine methyltransferase | - |
| CFSAN000624_RS24535 (CFSAN000624_024535) | 33912..34184 | + | 273 | Protein_35 | IS1 family transposase | - |
| CFSAN000624_RS26585 | 34204..34401 | - | 198 | WP_000844102 | hypothetical protein | - |
| CFSAN000624_RS24540 (CFSAN000624_024540) | 34406..34915 | - | 510 | WP_000893479 | restriction endonuclease | - |
| CFSAN000624_RS24545 (CFSAN000624_024545) | 35103..35414 | - | 312 | WP_001452736 | hypothetical protein | - |
| CFSAN000624_RS24550 (CFSAN000624_024550) | 35450..35764 | - | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| CFSAN000624_RS24555 (CFSAN000624_024555) | 35727..36104 | - | 378 | WP_000504251 | hypothetical protein | - |
| CFSAN000624_RS24560 (CFSAN000624_024560) | 36120..36425 | - | 306 | WP_000960954 | H-NS family nucleoid-associated regulatory protein | - |
| CFSAN000624_RS24565 (CFSAN000624_024565) | 36534..37268 | + | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| CFSAN000624_RS24570 (CFSAN000624_024570) | 37277..37558 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| CFSAN000624_RS24575 (CFSAN000624_024575) | 37568..37861 | + | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| CFSAN000624_RS24580 (CFSAN000624_024580) | 37911..38228 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| CFSAN000624_RS24585 (CFSAN000624_024585) | 38228..40828 | + | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| CFSAN000624_RS24590 (CFSAN000624_024590) | 40846..41559 | + | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| CFSAN000624_RS24595 (CFSAN000624_024595) | 41567..41794 | + | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| CFSAN000624_RS24600 (CFSAN000624_024600) | 41810..42850 | + | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| CFSAN000624_RS27685 (CFSAN000624_024605) | 42951..43079 | + | 129 | WP_071882546 | conjugal transfer protein TraN | - |
| CFSAN000624_RS24610 (CFSAN000624_024610) | 43069..43767 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| CFSAN000624_RS24615 (CFSAN000624_024615) | 43778..44662 | + | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| CFSAN000624_RS24620 (CFSAN000624_024620) | 44662..45822 | + | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| CFSAN000624_RS24625 (CFSAN000624_024625) | 45864..46859 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| CFSAN000624_RS24630 (CFSAN000624_024630) | 46859..47392 | + | 534 | WP_000731969 | phospholipase D family protein | - |
Host bacterium
| ID | 1316 | GenBank | NZ_CP017725 |
| Plasmid name | pSARB26_02 | Incompatibility group | IncN |
| Plasmid size | 49762 bp | Coordinate of oriT [Strand] | 15892..16540 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Stanleyville str. CFSAN000624 strain SGSC 2518 isolate SARB61 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |