Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100849 |
| Name | oriT_pRgalIE4872c |
| Organism | Rhizobium gallicum strain IE4872 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP017104 (483359..483400 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 20..25, 30..35 (CGTCGC..GCGACG) |
| Location of nic site | 9..10 |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 42 nt
>oriT_pRgalIE4872c
ATCCAAGGGCGCAATTATACGTCGCTGATGCGACGCCCTTGC
ATCCAAGGGCGCAATTATACGTCGCTGATGCGACGCCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file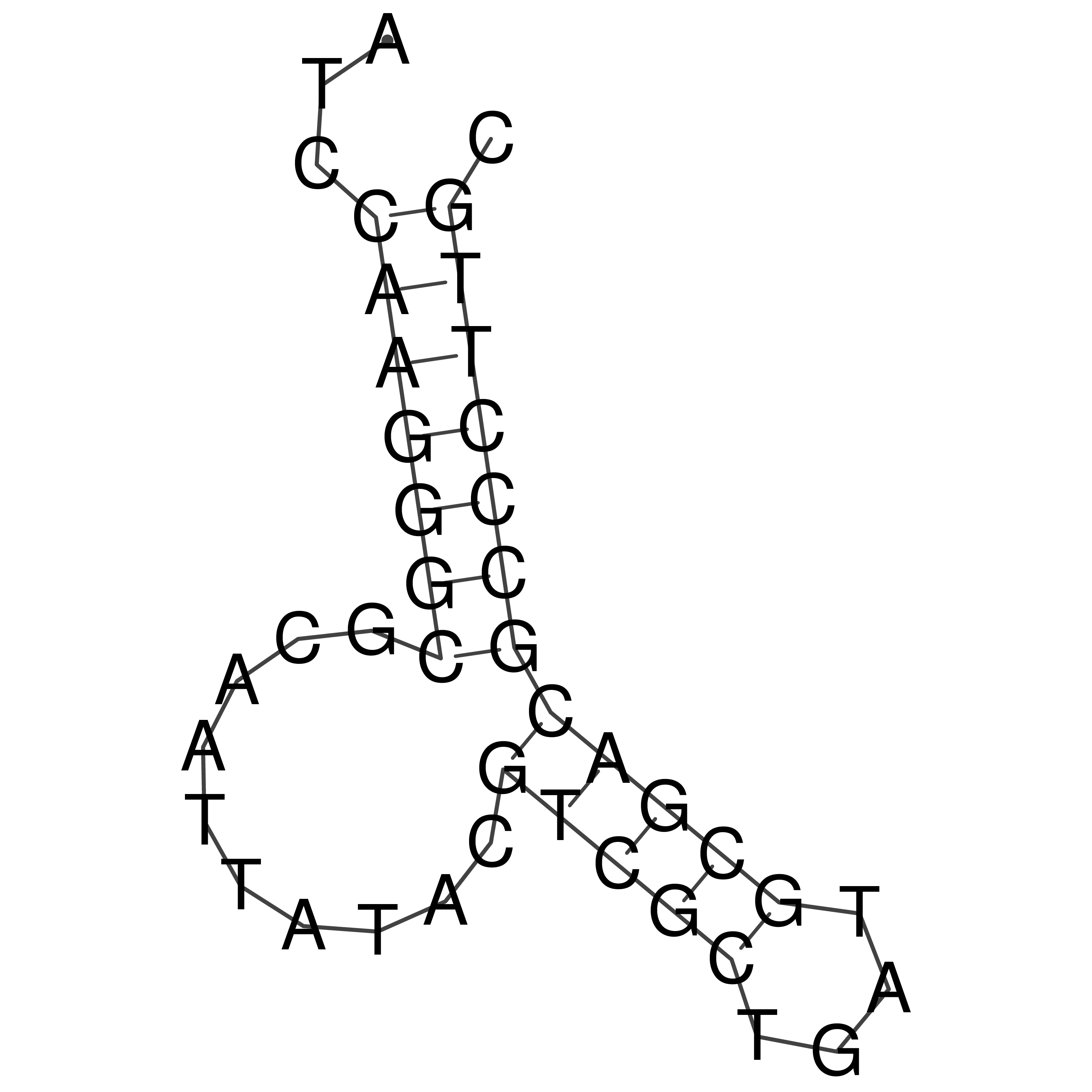
Relaxase
| ID | 921 | GenBank | WP_074070971 |
| Name | TraA_pRgalIE4872c |
UniProt ID | A0A1L5NRI8 |
| Length | 1103 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122305.53 Da Isoelectric Point: 9.8704
>WP_074070971.1 Ti-type conjugative transfer relaxase TraA [Rhizobium gallicum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKQGLAHEEFVIPADAPDWLRTIIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPVELTYEQNIALVREFVERHIIATGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGGKKVAVLGPDGNPMRNDSGKIVYELWAGSTDDFNALRDGWFVCQNRHLALAG
LDICIDGRSFEKQGIELTPTIHLGVGTTAIERKSEAATKPLTLERIELQEDRRAENARRVQRRPEIVLDL
ITREKSVFDERDVAKILHRYIDDADLFQSLLVRTLQSPETLQLEHERIEFATGVRVSGKYTTREVIQLEA
EMANRAIWLSHRSSHGVREAVLEATFSRHSRLSEEQKIAIAHVAGGERIAAVIGRAGAGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWVLRWNAGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRKQWMRDASLDLARGKVGKAVDAYR
AYGHFKGSELKVQAVENLIADWNGDCDPVKTTLILAHLRRDVRMLNEMARAKLVERGILAEGFVFKTEDG
NRKFAAGDQIVFLKNEGSLSVKNGMLAKVVEALPGRIVAEIGEGEHRRQVTVEQRFYNHVDHGYATTIHK
SQGATIDRVYILASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEKGAFY
KQALRFAEARGLHLVNVARTLVRDRLEWTVRQKQKLADLATRLAAIGGKLGLSTGAKKSPIQNIIKEAKP
MVSGIATFAKSLEQAVEDKVAADPGLKKQWEEVSSRFHLVYAQPETAFKAVNVDAILKDEAAAKSTIARI
GTQPDSFGALKGKTGILASRADKQGRERALNNAPALARKLERYLRQRADAERKHEVEERAVRLKVSVDIP
ALSPAAKRTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFARAVSERFGERTFLPLAAKDTSGQAFQT
VTTGMSAGQKAEVQSAWNSMRTVQQFAAHERTTETLKQVETLRQTKAQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKQGLAHEEFVIPADAPDWLRTIIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPVELTYEQNIALVREFVERHIIATGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGGKKVAVLGPDGNPMRNDSGKIVYELWAGSTDDFNALRDGWFVCQNRHLALAG
LDICIDGRSFEKQGIELTPTIHLGVGTTAIERKSEAATKPLTLERIELQEDRRAENARRVQRRPEIVLDL
ITREKSVFDERDVAKILHRYIDDADLFQSLLVRTLQSPETLQLEHERIEFATGVRVSGKYTTREVIQLEA
EMANRAIWLSHRSSHGVREAVLEATFSRHSRLSEEQKIAIAHVAGGERIAAVIGRAGAGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWVLRWNAGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRKQWMRDASLDLARGKVGKAVDAYR
AYGHFKGSELKVQAVENLIADWNGDCDPVKTTLILAHLRRDVRMLNEMARAKLVERGILAEGFVFKTEDG
NRKFAAGDQIVFLKNEGSLSVKNGMLAKVVEALPGRIVAEIGEGEHRRQVTVEQRFYNHVDHGYATTIHK
SQGATIDRVYILASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYEKGAFY
KQALRFAEARGLHLVNVARTLVRDRLEWTVRQKQKLADLATRLAAIGGKLGLSTGAKKSPIQNIIKEAKP
MVSGIATFAKSLEQAVEDKVAADPGLKKQWEEVSSRFHLVYAQPETAFKAVNVDAILKDEAAAKSTIARI
GTQPDSFGALKGKTGILASRADKQGRERALNNAPALARKLERYLRQRADAERKHEVEERAVRLKVSVDIP
ALSPAAKRTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFARAVSERFGERTFLPLAAKDTSGQAFQT
VTTGMSAGQKAEVQSAWNSMRTVQQFAAHERTTETLKQVETLRQTKAQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1L5NRI8 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 488517..501655
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IE4872_RS24990 (IE4872_PC00507) | 486841..487347 | + | 507 | WP_237361948 | conjugative transfer signal peptidase TraF | - |
| IE4872_RS24995 (IE4872_PC00508) | 487337..488500 | + | 1164 | WP_074070973 | conjugal transfer protein TraB | - |
| IE4872_RS25000 (IE4872_PC00509) | 488517..489131 | + | 615 | WP_074070974 | TraH family protein | virB1 |
| IE4872_RS25005 (IE4872_PC00510) | 489152..489529 | - | 378 | WP_074070975 | DUF5615 family PIN-like protein | - |
| IE4872_RS25010 (IE4872_PC00511) | 489526..490158 | - | 633 | WP_074070976 | DUF433 domain-containing protein | - |
| IE4872_RS25015 (IE4872_PC00512) | 490318..490533 | - | 216 | WP_040114408 | helix-turn-helix transcriptional regulator | - |
| IE4872_RS25020 (IE4872_PC00513) | 490751..491074 | + | 324 | WP_074070977 | transcriptional repressor TraM | - |
| IE4872_RS25025 (IE4872_PC00514) | 491078..491782 | - | 705 | WP_074071031 | autoinducer binding domain-containing protein | - |
| IE4872_RS25030 (IE4872_PC00515) | 492137..493435 | - | 1299 | WP_074070978 | IncP-type conjugal transfer protein TrbI | virB10 |
| IE4872_RS25035 (IE4872_PC00516) | 493447..493884 | - | 438 | WP_074070979 | conjugal transfer protein TrbH | - |
| IE4872_RS25040 (IE4872_PC00517) | 493888..494700 | - | 813 | WP_074070980 | P-type conjugative transfer protein TrbG | virB9 |
| IE4872_RS25045 (IE4872_PC00518) | 494718..495380 | - | 663 | WP_074070981 | conjugal transfer protein TrbF | virB8 |
| IE4872_RS25050 (IE4872_PC00519) | 495399..496580 | - | 1182 | WP_074070982 | P-type conjugative transfer protein TrbL | virB6 |
| IE4872_RS25055 (IE4872_PC00520) | 496574..496774 | - | 201 | WP_074070983 | entry exclusion protein TrbK | - |
| IE4872_RS25060 (IE4872_PC00521) | 496771..497574 | - | 804 | WP_074070984 | P-type conjugative transfer protein TrbJ | virB5 |
| IE4872_RS25065 (IE4872_PC00522) | 497546..500002 | - | 2457 | WP_074070985 | conjugal transfer protein TrbE | virb4 |
| IE4872_RS25070 (IE4872_PC00523) | 500013..500312 | - | 300 | WP_074070986 | conjugal transfer protein TrbD | virB3 |
| IE4872_RS25075 (IE4872_PC00524) | 500305..500688 | - | 384 | WP_074070987 | TrbC/VirB2 family protein | virB2 |
| IE4872_RS25080 (IE4872_PC00525) | 500678..501655 | - | 978 | WP_074070988 | P-type conjugative transfer ATPase TrbB | virB11 |
| IE4872_RS25085 (IE4872_PC00526) | 501666..502289 | - | 624 | WP_074070989 | acyl-homoserine-lactone synthase TraI | - |
| IE4872_RS25090 (IE4872_PC00528) | 502611..503894 | + | 1284 | WP_074070990 | plasmid partitioning protein RepA | - |
| IE4872_RS25095 (IE4872_PC00529) | 503891..504922 | + | 1032 | WP_074070991 | plasmid partitioning protein RepB | - |
| IE4872_RS25100 (IE4872_PC00530) | 505117..506328 | + | 1212 | WP_074070992 | plasmid replication protein RepC | - |
Host bacterium
| ID | 1309 | GenBank | NZ_CP017104 |
| Plasmid name | pRgalIE4872c | Incompatibility group | _ |
| Plasmid size | 506339 bp | Coordinate of oriT [Strand] | 483359..483400 [+] |
| Host baterium | Rhizobium gallicum strain IE4872 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nodA, nodE, nodM, fixN, fixO, fixQ, fixS, nodB, nodC, nodS, nodU, nodI, nodJ, nodH, nifT, nifZ, nifB, nifA, fixX, fixC, fixB, fixA, nifW, nifS, nifH, nifD, nifK, nifE, nifX, mLTONO_5203 |
| Anti-CRISPR | AcrIIC1 |