Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100809 |
| Name | oriT_pSLy2 |
| Organism | Escherichia coli strain 210205630 isolate 210205630 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP015914 (42515..42804 [+], 290 nt) |
| oriT length | 290 nt |
| IRs (inverted repeats) | 225..232, 235..242 n (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 251..252 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 290 nt
>oriT_pSLy2
CGCACCGCTAGCAGCGCCCCTAGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCGCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAACTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
CGCACCGCTAGCAGCGCCCCTAGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCGCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAACTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file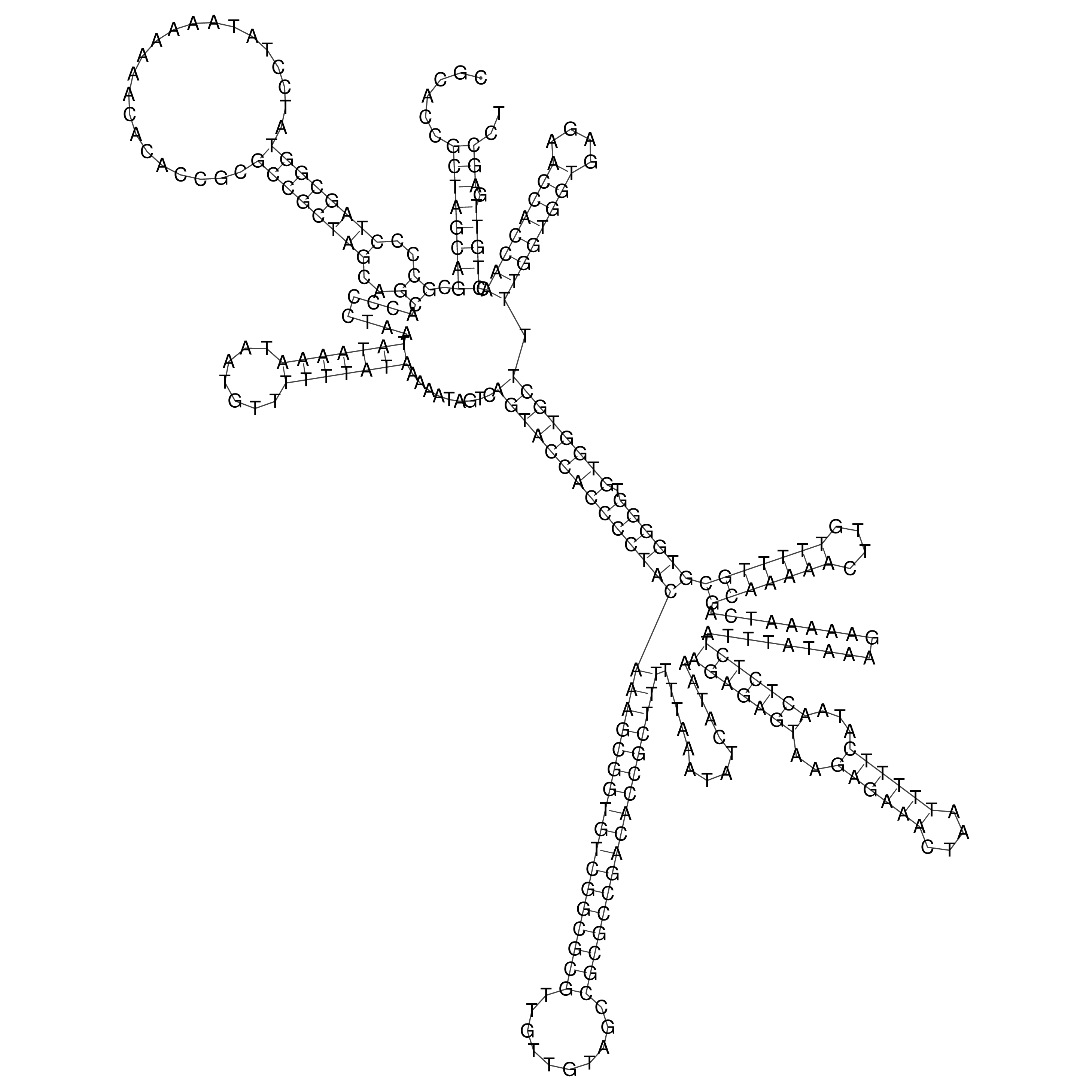
Relaxase
| ID | 881 | GenBank | WP_054377139 |
| Name | TraI_pSLy2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191884.75 Da Isoelectric Point: 6.1058
>WP_054377139.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHTVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNSYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVTSVSEDAMTVVVPGRAEPASLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGEILLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFTDLGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESGRPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGAGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSGRQQTRVIRPAQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGSRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETVRRADEIVRKMAENKPDLP
DSKTEQAVREIAGQERDRSAISEREAALPESVLRESQREREAVREIARENLLQERLQQMERDMVRDLQKE
KTLGGD
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHTVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNSYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVTSVSEDAMTVVVPGRAEPASLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGEILLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFTDLGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESGRPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGAGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSGRQQTRVIRPAQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGSRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETVRRADEIVRKMAENKPDLP
DSKTEQAVREIAGQERDRSAISEREAALPESVLRESQREREAVREIARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 18125..43366
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A9C00_RS25330 (A9C00_25330) | 18125..20287 | - | 2163 | WP_065304458 | type IV conjugative transfer system coupling protein TraD | virb4 |
| A9C00_RS25335 (A9C00_25335) | 20540..21271 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| A9C00_RS25340 (A9C00_25340) | 21303..21806 | - | 504 | WP_000605510 | hypothetical protein | - |
| A9C00_RS25345 (A9C00_25345) | 21821..24643 | - | 2823 | WP_001007048 | conjugal transfer mating-pair stabilization protein TraG | traG |
| A9C00_RS25350 (A9C00_25350) | 24640..26013 | - | 1374 | WP_170926237 | conjugal transfer pilus assembly protein TraH | traH |
| A9C00_RS25355 (A9C00_25355) | 26013..26375 | - | 363 | WP_001443292 | P-type conjugative transfer protein TrbJ | - |
| A9C00_RS25360 (A9C00_25360) | 26305..26850 | - | 546 | WP_000052613 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| A9C00_RS25365 (A9C00_25365) | 26837..27073 | - | 237 | WP_054377141 | type-F conjugative transfer system pilin chaperone TraQ | - |
| A9C00_RS25370 (A9C00_25370) | 27234..27977 | - | 744 | WP_039002205 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| A9C00_RS25375 (A9C00_25375) | 27970..28230 | - | 261 | WP_000864348 | conjugal transfer protein TrbE | - |
| A9C00_RS25380 (A9C00_25380) | 28257..30065 | - | 1809 | WP_000821844 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| A9C00_RS25385 (A9C00_25385) | 30062..30700 | - | 639 | WP_000777691 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| A9C00_RS25390 (A9C00_25390) | 30709..31701 | - | 993 | WP_023148235 | conjugal transfer pilus assembly protein TraU | traU |
| A9C00_RS25395 (A9C00_25395) | 31698..32330 | - | 633 | WP_001749440 | type-F conjugative transfer system protein TraW | traW |
| A9C00_RS25400 (A9C00_25400) | 32327..32713 | - | 387 | WP_039002198 | type-F conjugative transfer system protein TrbI | - |
| A9C00_RS25405 (A9C00_25405) | 32710..35337 | - | 2628 | WP_001064265 | type IV secretion system protein TraC | virb4 |
| A9C00_RS28955 | 35497..35718 | - | 222 | WP_001278692 | conjugal transfer protein TraR | - |
| A9C00_RS25410 (A9C00_25410) | 35853..36368 | - | 516 | WP_000724019 | type IV conjugative transfer system lipoprotein TraV | traV |
| A9C00_RS25415 (A9C00_25415) | 36365..36685 | - | 321 | WP_001057285 | conjugal transfer protein TrbD | - |
| A9C00_RS25420 (A9C00_25420) | 36672..37259 | - | 588 | WP_000002772 | conjugal transfer pilus-stabilizing protein TraP | - |
| A9C00_RS25425 (A9C00_25425) | 37249..38676 | - | 1428 | WP_000146690 | F-type conjugal transfer pilus assembly protein TraB | traB |
| A9C00_RS25430 (A9C00_25430) | 38676..39404 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| A9C00_RS25435 (A9C00_25435) | 39391..39957 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| A9C00_RS25440 (A9C00_25440) | 39979..40290 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| A9C00_RS25445 (A9C00_25445) | 40295..40657 | - | 363 | WP_000994784 | type IV conjugative transfer system pilin TraA | - |
| A9C00_RS25450 (A9C00_25450) | 40690..41085 | - | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| A9C00_RS25455 (A9C00_25455) | 41184..41873 | - | 690 | WP_039002187 | conjugal transfer transcriptional regulator TraJ | - |
| A9C00_RS25460 (A9C00_25460) | 42060..42443 | - | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| A9C00_RS25465 (A9C00_25465) | 42776..43366 | + | 591 | WP_001376243 | transglycosylase SLT domain-containing protein | virB1 |
| A9C00_RS25470 (A9C00_25470) | 43663..44484 | - | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| A9C00_RS31415 | 44606..44788 | - | 183 | WP_170979925 | hypothetical protein | - |
| A9C00_RS25480 (A9C00_25480) | 44894..47860 | - | 2967 | WP_000008826 | Tn3 family transposase | - |
Host bacterium
| ID | 1269 | GenBank | NZ_CP015914 |
| Plasmid name | pSLy2 | Incompatibility group | IncFIC |
| Plasmid size | 129035 bp | Coordinate of oriT [Strand] | 42515..42804 [+] |
| Host baterium | Escherichia coli strain 210205630 isolate 210205630 |
Cargo genes
| Drug resistance gene | oqxB, oqxA, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |