Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100804 |
| Name | oriT_pSKGH01-5 |
| Organism | Klebsiella pneumoniae strain SKGH01 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP015505 (12479..12953 [-], 475 nt) |
| oriT length | 475 nt |
| IRs (inverted repeats) | IR1: 145..155, 161..171 (ATGTGTAATAC..GTATTACACAT) IR2: 208..218, 223..233 (GCATTTACGAT..ATCGTAATTGC) IR3: 335..344, 347..356 (CCTTTCATGC..GCATGAAAGG) |
| Location of nic site | 242..243 |
| Conserved sequence flanking the nic site |
GGTGT|ATAGC |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 475 nt
>oriT_pSKGH01-5
ACTCCGATACGTTGCCCTCCGAACGGTATTCAAGGTCGATTTTTTGCGCTTCGGTCACTCTTAAACTGATAGATGGCATAGGTTTCCTTTGTGTAATACCGATGTAATACATACGAATCTAGCATATATGCGGCTTAATTACACATGTGTAATACGTTGTGTATTACACATTAAAACACAAATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGGGAGAAACCGGTGTTACGTGCAAGTGAATCGCTCAAAAAGCGTTCACATTCACACCTTTCATGCTTGCATGAAAGGAAACGGACGGGAATTAGACAAAAATAAGACACGAGGAGTAAGTTATTGAGACAAGAAAAGGACACAAATAAGACATTTTTTAGAAAAAAACATTGACTTGAGACTAGAAATGGACAATA
ACTCCGATACGTTGCCCTCCGAACGGTATTCAAGGTCGATTTTTTGCGCTTCGGTCACTCTTAAACTGATAGATGGCATAGGTTTCCTTTGTGTAATACCGATGTAATACATACGAATCTAGCATATATGCGGCTTAATTACACATGTGTAATACGTTGTGTATTACACATTAAAACACAAATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATACCATACCACGCCTTTTTTAAGGGAGAAACCGGTGTTACGTGCAAGTGAATCGCTCAAAAAGCGTTCACATTCACACCTTTCATGCTTGCATGAAAGGAAACGGACGGGAATTAGACAAAAATAAGACACGAGGAGTAAGTTATTGAGACAAGAAAAGGACACAAATAAGACATTTTTTAGAAAAAAACATTGACTTGAGACTAGAAATGGACAATA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file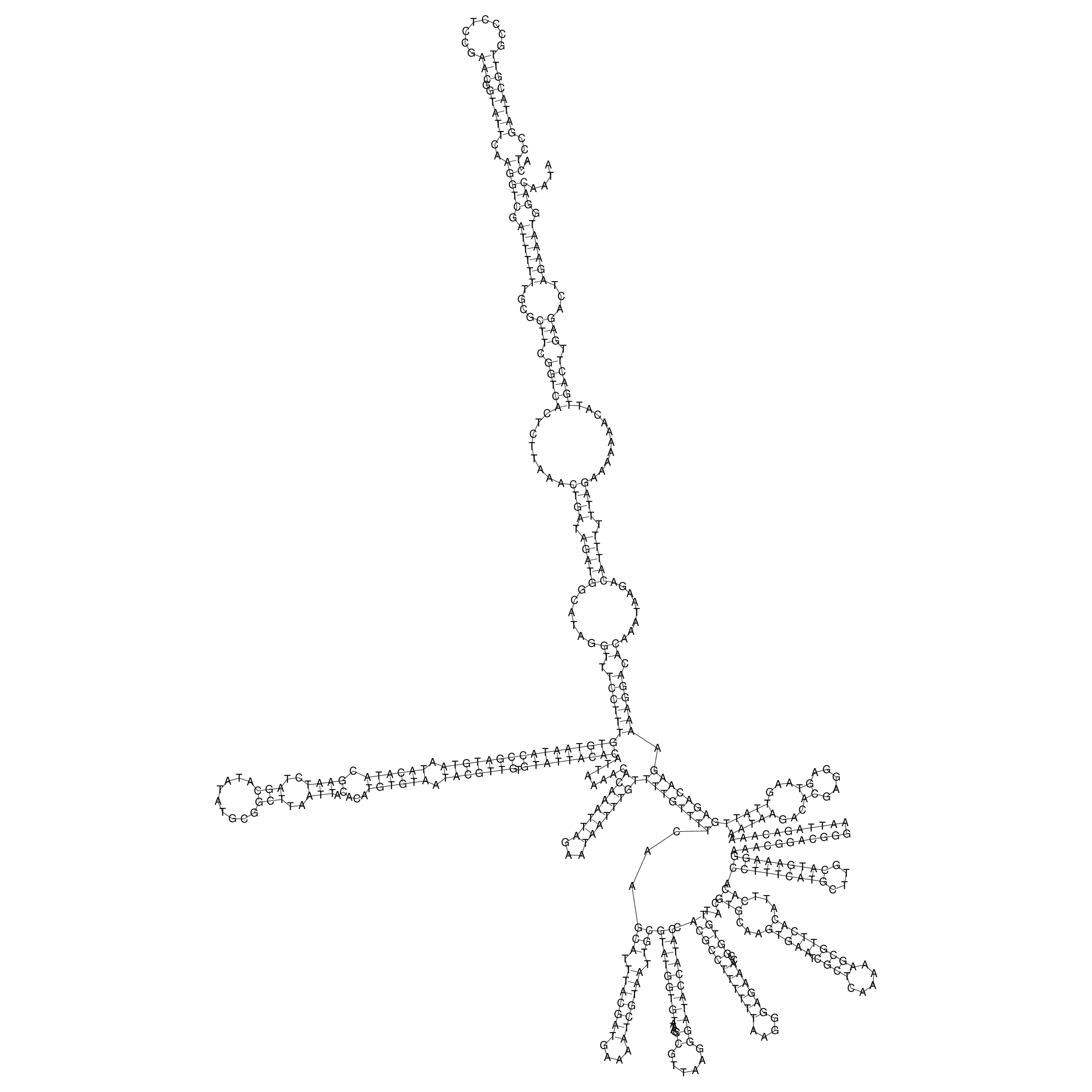
Relaxase
| ID | 876 | GenBank | WP_000932317 |
| Name | Relaxase_pSKGH01-5 |
UniProt ID | _ |
| Length | 969 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 969 a.a. Molecular weight: 108425.51 Da Isoelectric Point: 9.7925
>WP_000932317.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYRRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYRRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13215..31003
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A7B01_RS29985 (A7B01_29995) | 8468..8755 | + | 288 | WP_001098466 | hypothetical protein | - |
| A7B01_RS29990 (A7B01_30000) | 8810..9052 | + | 243 | WP_000526841 | hypothetical protein | - |
| A7B01_RS33785 | 9205..9360 | + | 156 | WP_021536821 | hypothetical protein | - |
| A7B01_RS29995 (A7B01_30005) | 9433..9690 | + | 258 | WP_000334833 | hypothetical protein | - |
| A7B01_RS30000 (A7B01_30010) | 9695..9883 | + | 189 | WP_001001010 | hypothetical protein | - |
| A7B01_RS30005 (A7B01_30015) | 9923..10693 | - | 771 | WP_000586559 | SprT-like domain-containing protein | - |
| A7B01_RS30010 (A7B01_30020) | 10907..11275 | - | 369 | WP_000414914 | plasmid stabilization protein StbC | - |
| A7B01_RS30015 (A7B01_30025) | 11277..11987 | - | 711 | WP_000686338 | StbB family protein | - |
| A7B01_RS30020 (A7B01_30030) | 12000..12443 | - | 444 | WP_024249956 | hypothetical protein | - |
| A7B01_RS30025 (A7B01_30035) | 12875..13222 | + | 348 | WP_001139329 | hypothetical protein | - |
| A7B01_RS30030 (A7B01_30040) | 13215..14792 | + | 1578 | WP_001001035 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| A7B01_RS30035 (A7B01_30045) | 14795..17704 | + | 2910 | WP_000932317 | MobF family relaxase | - |
| A7B01_RS30040 (A7B01_30050) | 17981..18175 | - | 195 | WP_000529710 | hypothetical protein | - |
| A7B01_RS30045 (A7B01_30055) | 18187..18669 | - | 483 | WP_036896345 | GNAT family protein | - |
| A7B01_RS30050 (A7B01_30060) | 18682..19236 | - | 555 | WP_000616063 | phospholipase D family protein | - |
| A7B01_RS30055 (A7B01_30065) | 19199..20230 | - | 1032 | WP_000138004 | P-type DNA transfer ATPase VirB11 | virB11 |
| A7B01_RS30060 (A7B01_30070) | 20223..21377 | - | 1155 | WP_001224296 | type IV secretion system protein VirB10 | virB10 |
| A7B01_RS30065 (A7B01_30075) | 21374..22261 | - | 888 | WP_000744355 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| A7B01_RS30070 (A7B01_30080) | 22264..22980 | - | 717 | WP_000914645 | type IV secretion system protein | virB8 |
| A7B01_RS33790 | 22983..23153 | - | 171 | WP_021536809 | hypothetical protein | - |
| A7B01_RS30075 (A7B01_30085) | 23248..24291 | - | 1044 | WP_000885742 | type IV secretion system protein | virB6 |
| A7B01_RS30080 (A7B01_30090) | 24310..24546 | - | 237 | WP_000733633 | EexN family lipoprotein | - |
| A7B01_RS30085 (A7B01_30095) | 24550..25257 | - | 708 | WP_036896352 | type IV secretion system protein | virB5 |
| A7B01_RS30090 (A7B01_30100) | 25267..27897 | - | 2631 | WP_000257330 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| A7B01_RS30095 (A7B01_30105) | 27897..28211 | - | 315 | WP_001053888 | VirB3 family type IV secretion system protein | virB3 |
| A7B01_RS30100 (A7B01_30110) | 28261..28551 | - | 291 | WP_001090269 | TrbC/VirB2 family protein | virB2 |
| A7B01_RS30105 (A7B01_30115) | 28535..28852 | - | 318 | WP_001267114 | hypothetical protein | - |
| A7B01_RS30110 (A7B01_30120) | 28875..29615 | - | 741 | WP_000815895 | lytic transglycosylase domain-containing protein | virB1 |
| A7B01_RS32425 | 29678..29914 | + | 237 | WP_000089855 | H-NS histone family protein | - |
| A7B01_RS30120 (A7B01_30130) | 29926..30363 | + | 438 | WP_000034608 | hypothetical protein | - |
| A7B01_RS30125 (A7B01_30135) | 30369..30686 | + | 318 | WP_001226231 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| A7B01_RS30130 (A7B01_30140) | 30689..31003 | + | 315 | WP_000851068 | hypothetical protein | tfc15 |
| A7B01_RS30135 (A7B01_30145) | 30979..31704 | + | 726 | WP_223307254 | restriction endonuclease | - |
| A7B01_RS34420 | 31705..32106 | + | 402 | WP_023187076 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| A7B01_RS30150 (A7B01_30160) | 32166..32480 | - | 315 | WP_001280992 | hypothetical protein | - |
| A7B01_RS32050 | 32506..32832 | - | 327 | WP_000817647 | endoribonuclease MazF | - |
| A7B01_RS30155 (A7B01_30165) | 32822..33118 | - | 297 | WP_223307255 | hypothetical protein | - |
| A7B01_RS30160 (A7B01_30170) | 33088..33630 | - | 543 | WP_000711249 | hypothetical protein | - |
| A7B01_RS30165 (A7B01_30175) | 33630..33875 | - | 246 | WP_000195442 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
Host bacterium
| ID | 1264 | GenBank | NZ_CP015505 |
| Plasmid name | pSKGH01-5 | Incompatibility group | _ |
| Plasmid size | 34611 bp | Coordinate of oriT [Strand] | 12479..12953 [-] |
| Host baterium | Klebsiella pneumoniae strain SKGH01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |