Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100755 |
| Name | oriT_PDM02 |
| Organism | Salmonella enterica subsp. enterica serovar Anatum strain GT-01 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013221 (36168..36270 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | IR1: 19..26, 30..37 (GTCGGGGC..GCCCTGAC) IR2: 44..58, 67..81 (GCAATTGTATAATCG..CGGTTATACAATTGC) |
| Location of nic site | 89..90 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 103 nt
>oriT_PDM02
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file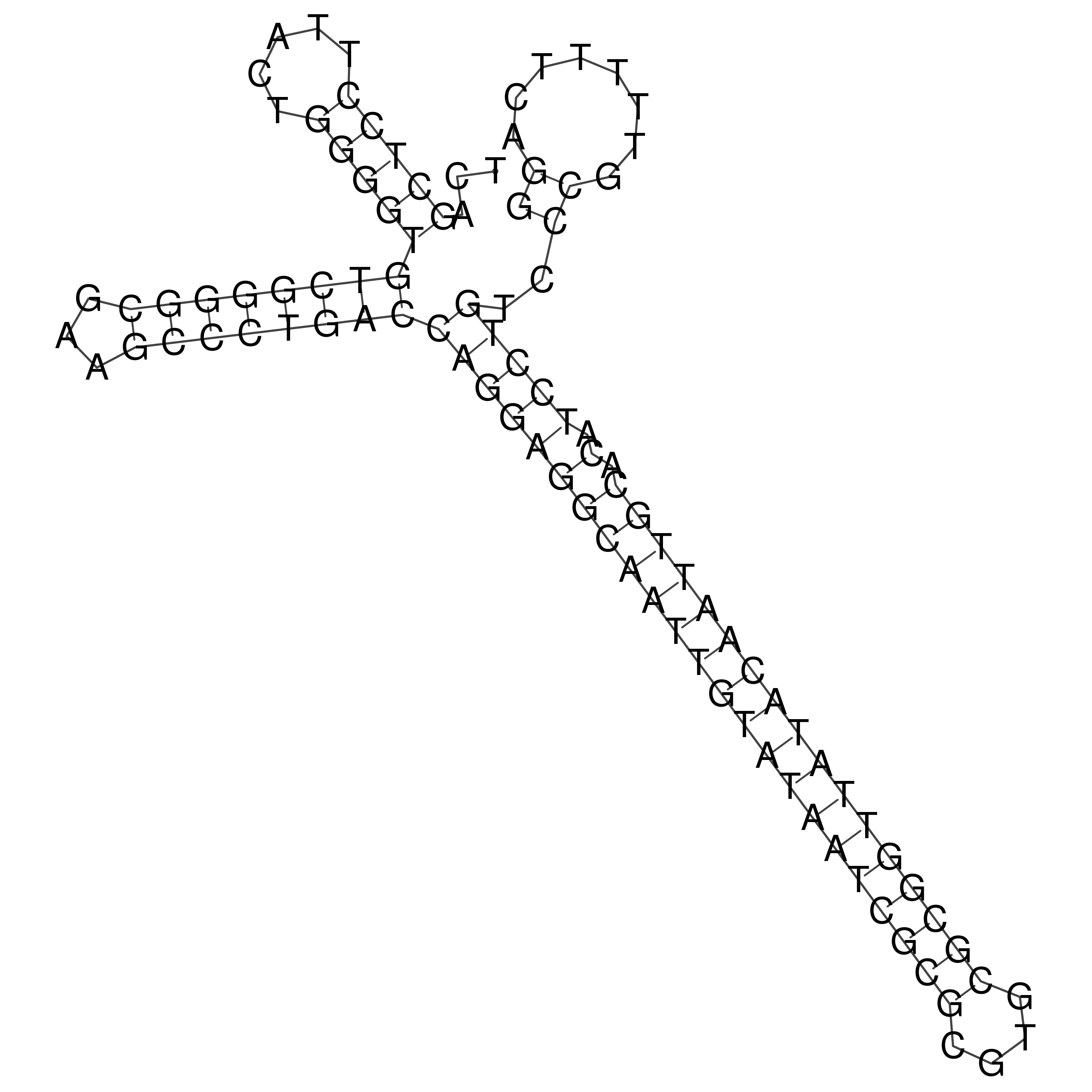
Relaxase
| ID | 827 | GenBank | WP_001354015 |
| Name | NikB_PDM02 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104018.47 Da Isoelectric Point: 7.3526
>WP_001354015.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCLAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPENRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1372..30802
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AS587_RS00010 (AS587_23695) | 67..1221 | + | 1155 | WP_001139957 | site-specific integrase | - |
| AS587_RS00015 (AS587_23700) | 1372..2196 | + | 825 | WP_001238939 | conjugal transfer protein TraE | traE |
| AS587_RS00020 (AS587_23705) | 2281..3483 | + | 1203 | WP_000976351 | conjugal transfer protein TraF | - |
| AS587_RS00025 (AS587_23710) | 3543..4127 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| AS587_RS00030 (AS587_23715) | 4522..4980 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| AS587_RS00035 (AS587_23720) | 4977..5795 | + | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| AS587_RS00040 (AS587_23725) | 5792..6940 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| AS587_RS24355 | 6937..7227 | + | 291 | WP_001299214 | hypothetical protein | traK |
| AS587_RS00045 (AS587_23730) | 7242..7793 | + | 552 | WP_000014583 | phospholipase D family protein | - |
| AS587_RS00050 (AS587_23735) | 7883..11650 | + | 3768 | WP_001141529 | LPD7 domain-containing protein | - |
| AS587_RS00055 (AS587_23740) | 11668..12015 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| AS587_RS00060 (AS587_23745) | 12012..12704 | + | 693 | WP_000138548 | DotI/IcmL family type IV secretion protein | traM |
| AS587_RS00065 (AS587_23750) | 12715..13698 | + | 984 | WP_001191877 | IncI1-type conjugal transfer protein TraN | traN |
| AS587_RS00070 (AS587_23755) | 13701..14990 | + | 1290 | WP_001272005 | conjugal transfer protein TraO | traO |
| AS587_RS00075 (AS587_23760) | 14990..15694 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| AS587_RS00080 (AS587_23765) | 15694..16221 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| AS587_RS00085 (AS587_23770) | 16272..16676 | + | 405 | WP_000086957 | IncI1-type conjugal transfer protein TraR | traR |
| AS587_RS00090 (AS587_23775) | 16740..16928 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| AS587_RS00095 (AS587_23780) | 16912..17712 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| AS587_RS00100 (AS587_23785) | 17802..20846 | + | 3045 | WP_001024780 | IncI1-type conjugal transfer protein TraU | traU |
| AS587_RS24360 | 20846..21460 | + | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| AS587_RS00105 (AS587_23790) | 21427..22629 | + | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| AS587_RS00110 (AS587_23795) | 22658..23242 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| AS587_RS00115 (AS587_23800) | 23339..25501 | + | 2163 | WP_000698351 | DotA/TraY family protein | traY |
| AS587_RS00120 (AS587_23805) | 25566..26228 | + | 663 | WP_000653334 | plasmid IncI1-type surface exclusion protein ExcA | - |
| AS587_RS00125 (AS587_23810) | 26300..26509 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| AS587_RS26545 | 26901..27077 | + | 177 | WP_001054904 | hypothetical protein | - |
| AS587_RS27035 | 27142..27237 | - | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| AS587_RS27040 | 27798..27989 | + | 192 | WP_174715448 | hypothetical protein | - |
| AS587_RS00135 (AS587_23820) | 28061..28213 | - | 153 | WP_001331364 | Hok/Gef family protein | - |
| AS587_RS00140 (AS587_23825) | 28505..29713 | + | 1209 | WP_000121274 | IncI1-type conjugal transfer protein TrbA | trbA |
| AS587_RS00145 (AS587_23830) | 29732..30802 | + | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| AS587_RS00150 (AS587_23835) | 30795..33086 | + | 2292 | WP_001289276 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 1215 | GenBank | NZ_CP013221 |
| Plasmid name | PDM02 | Incompatibility group | IncI1 |
| Plasmid size | 112176 bp | Coordinate of oriT [Strand] | 36168..36270 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Anatum strain GT-01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |