Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100750 |
| Name | oriT_p2009C-3133-2 |
| Organism | Escherichia coli strain 2009C-3133 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013026 (38763..39224 [+], 462 nt) |
| oriT length | 462 nt |
| IRs (inverted repeats) | 142..151, 154..163 (ATAGGGTCGT..ACGACCCTAT) 231..238, 241..248 (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 257..258 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 462 nt
>oriT_p2009C-3133-2
AAGAAGAGCACCCCTAGCAGCGCCCCTAGGGTCATACTATAAAAAAACGCACGGCGCTGCTAGGGGCGGCCCTAATATATCCAAATGTTTTTCATAAAATTTGTCAGTACTGACCCTAACAAGGGTCGCCATAGGGTCGCCATAGGGTCGTCAACGACCCTATTTAATAATGTAAAATAAATTAAAATACATTATTTAAAACATAAGCTAATGATTCAAAAAGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCTTTTTGTGGAGTGGGTTAAATTATTTACGGATAAAGTCACCAGAGGTGGAAAAATGAAAAAATGGATGTTAGCCATCTGCCTGATGTTTATAAATGAGATCTGCCATGCCACTGATTGCTTTGATCTTGCAGGCCGGGATTACAAAATAGATCCTGATTTACTGAGA
AAGAAGAGCACCCCTAGCAGCGCCCCTAGGGTCATACTATAAAAAAACGCACGGCGCTGCTAGGGGCGGCCCTAATATATCCAAATGTTTTTCATAAAATTTGTCAGTACTGACCCTAACAAGGGTCGCCATAGGGTCGCCATAGGGTCGTCAACGACCCTATTTAATAATGTAAAATAAATTAAAATACATTATTTAAAACATAAGCTAATGATTCAAAAAGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCTTTTTGTGGAGTGGGTTAAATTATTTACGGATAAAGTCACCAGAGGTGGAAAAATGAAAAAATGGATGTTAGCCATCTGCCTGATGTTTATAAATGAGATCTGCCATGCCACTGATTGCTTTGATCTTGCAGGCCGGGATTACAAAATAGATCCTGATTTACTGAGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file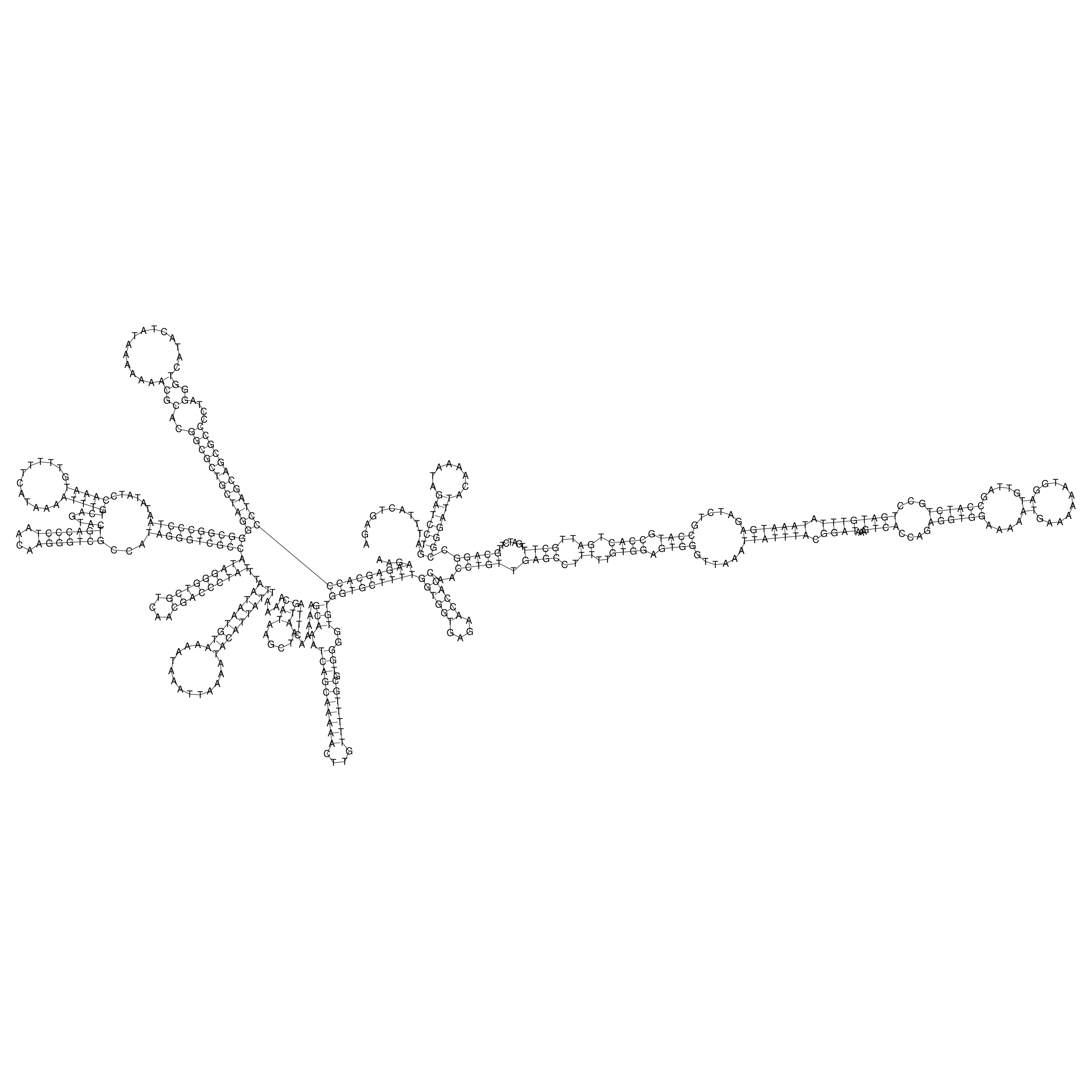
Relaxase
| ID | 822 | GenBank | WP_060565186 |
| Name | TraI_p2009C-3133-2 |
UniProt ID | A0A0P0T0S7 |
| Length | 1756 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192318.04 Da Isoelectric Point: 5.9386
>WP_060565186.1 conjugative transfer relaxase/helicase TraI [Escherichia coli]
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEIKMAEWMQT
LKETGFDIRAYRDAADQRADLRTQTPRPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDTGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKTGEESVAQVSGVREQAILTQAIRSELKTQDVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVTERVTAQSNSLTLRNAQGETRVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAELASLPVADSPFTALKL
ESGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFTELGGEIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEMLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISRDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARQLQDVAAGRAVLRQAGLARGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQEGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAGLERERAVTSEREAALPESVLREPQREREAVREVVRENLLQERLQQMERDMVRDLQKE
KTLGGD
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEIKMAEWMQT
LKETGFDIRAYRDAADQRADLRTQTPRPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDTGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKTGEESVAQVSGVREQAILTQAIRSELKTQDVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVTERVTAQSNSLTLRNAQGETRVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAELASLPVADSPFTALKL
ESGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSQVDLLTEAKSFAAEGTSFTELGGEIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEMLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISRDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARQLQDVAAGRAVLRQAGLARGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQEGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGRTEQAVREIAGLERERAVTSEREAALPESVLREPQREREAVREVVRENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13403..39620
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MJ49_RS26605 (MJ49_26570) | 13403..15574 | - | 2172 | WP_060565188 | type IV conjugative transfer system coupling protein TraD | virb4 |
| MJ49_RS26610 (MJ49_26575) | 15827..16558 | - | 732 | WP_000850422 | conjugal transfer complement resistance protein TraT | - |
| MJ49_RS26615 (MJ49_26580) | 16590..17093 | - | 504 | WP_060565200 | surface exclusion protein | - |
| MJ49_RS26620 (MJ49_26585) | 17108..19930 | - | 2823 | WP_054622909 | conjugal transfer mating-pair stabilization protein TraG | traG |
| MJ49_RS26625 (MJ49_26590) | 19927..21300 | - | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| MJ49_RS26630 (MJ49_26595) | 21300..21662 | - | 363 | WP_023181029 | P-type conjugative transfer protein TrbJ | - |
| MJ49_RS26635 (MJ49_26600) | 21592..22137 | - | 546 | WP_000052614 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| MJ49_RS26640 (MJ49_26605) | 22124..22408 | - | 285 | WP_023181030 | type-F conjugative transfer system pilin chaperone TraQ | - |
| MJ49_RS26650 (MJ49_26615) | 22807..23148 | - | 342 | WP_023181031 | conjugal transfer protein TrbA | - |
| MJ49_RS26655 (MJ49_26620) | 23162..23905 | - | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| MJ49_RS26660 (MJ49_26625) | 23898..24155 | - | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| MJ49_RS26665 (MJ49_26630) | 24182..25990 | - | 1809 | WP_060565190 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| MJ49_RS26670 (MJ49_26635) | 25987..26625 | - | 639 | WP_000777692 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| MJ49_RS26675 (MJ49_26640) | 26634..27626 | - | 993 | WP_001712363 | conjugal transfer pilus assembly protein TraU | traU |
| MJ49_RS26680 (MJ49_26645) | 27623..28255 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| MJ49_RS26685 (MJ49_26650) | 28252..28638 | - | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| MJ49_RS26690 (MJ49_26655) | 28635..31265 | - | 2631 | WP_044869719 | type IV secretion system protein TraC | virb4 |
| MJ49_RS30085 | 31658..31837 | - | 180 | WP_071887283 | conjugal transfer protein TraR | - |
| MJ49_RS33230 | 32014..32199 | - | 186 | WP_001444126 | type IV conjugative transfer system lipoprotein TraV | traV |
| MJ49_RS26695 (MJ49_26660) | 32132..32530 | - | 399 | WP_236927495 | type IV conjugative transfer system lipoprotein TraV | traV |
| MJ49_RS30090 | 32527..32779 | - | 253 | Protein_37 | conjugal transfer protein TrbG | - |
| MJ49_RS31965 | 32772..33098 | - | 327 | Protein_38 | conjugal transfer protein TrbD | - |
| MJ49_RS26700 (MJ49_26665) | 33085..33681 | - | 597 | WP_077870409 | conjugal transfer pilus-stabilizing protein TraP | - |
| MJ49_RS26705 (MJ49_26670) | 33659..35100 | - | 1442 | Protein_40 | F-type conjugal transfer pilus assembly protein TraB | - |
| MJ49_RS26710 (MJ49_26675) | 35100..35828 | - | 729 | WP_001230788 | type-F conjugative transfer system secretin TraK | traK |
| MJ49_RS26715 (MJ49_26680) | 35815..36381 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| MJ49_RS26720 (MJ49_26685) | 36403..36714 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| MJ49_RS26725 (MJ49_26690) | 36719..37081 | - | 363 | WP_000340279 | type IV conjugative transfer system pilin TraA | - |
| MJ49_RS26730 (MJ49_26695) | 37115..37342 | - | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| MJ49_RS26735 (MJ49_26700) | 37436..38122 | - | 687 | WP_000332484 | PAS domain-containing protein | - |
| MJ49_RS26740 (MJ49_26705) | 38313..38696 | - | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| MJ49_RS26745 (MJ49_26710) | 38973..39620 | + | 648 | WP_000614934 | transglycosylase SLT domain-containing protein | virB1 |
| MJ49_RS26750 (MJ49_26715) | 39917..40738 | - | 822 | WP_021573168 | DUF932 domain-containing protein | - |
| MJ49_RS26755 (MJ49_26720) | 40855..41142 | - | 288 | WP_000107538 | hypothetical protein | - |
| MJ49_RS33235 | 41443..41604 | + | 162 | Protein_51 | hypothetical protein | - |
| MJ49_RS33240 | 41614..41844 | - | 231 | WP_001426396 | hypothetical protein | - |
| MJ49_RS26760 (MJ49_26725) | 42064..42189 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| MJ49_RS33245 | 42131..42280 | - | 150 | Protein_54 | plasmid maintenance protein Mok | - |
| MJ49_RS26770 (MJ49_26735) | 42459..43221 | - | 763 | Protein_55 | plasmid SOS inhibition protein A | - |
| MJ49_RS26775 (MJ49_26740) | 43218..43652 | - | 435 | WP_000845930 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 1210 | GenBank | NZ_CP013026 |
| Plasmid name | p2009C-3133-2 | Incompatibility group | IncFII |
| Plasmid size | 63800 bp | Coordinate of oriT [Strand] | 38763..39224 [+] |
| Host baterium | Escherichia coli strain 2009C-3133 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |