Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100745 |
| Name | oriT_p12-4374_62 |
| Organism | Salmonella enterica subsp. enterica serovar Heidelberg strain 12-4374 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012928 (36265..36554 [+], 290 nt) |
| oriT length | 290 nt |
| IRs (inverted repeats) | 225..232, 235..242 n (GCAAAAAC..GTTTTTGC) |
| Location of nic site | 251..252 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 290 nt
>oriT_p12-4374_62
CGCACCGCTAGCAGCGCCCCTGGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCGCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAGCTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTACTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
CGCACCGCTAGCAGCGCCCCTGGCGGTATCCTATAAAAAAACACACCGCGCCGCTAGCAGCACCCCTAATATAAAATAATGTTTTTTATAAAAATAGTCAGTACCACCCCTACAAAGCGGTGTCGGCGCGTTGTTGTAGCCGCGCCGACACCGCTTTTTTAAATATCATAAAGAGAGTAAGAGAAGCTAATTTTTCATAACTCTCTATTTATAAAGAAAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTACTTTTGGTGGTGAGAACCACCAACCTGTTGAGCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file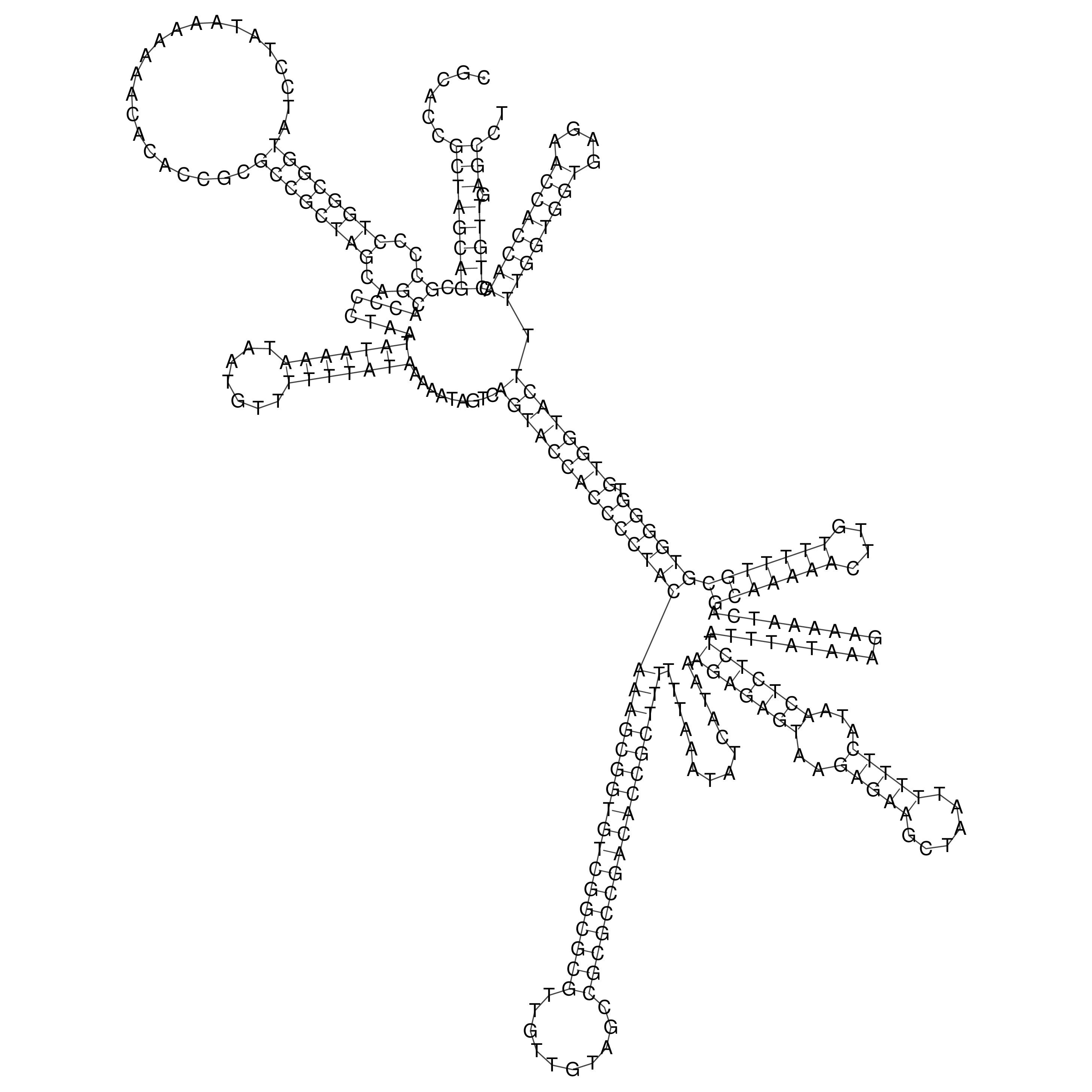
Relaxase
| ID | 817 | GenBank | WP_044864167 |
| Name | TraI_p12-4374_62 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191851.73 Da Isoelectric Point: 5.8723
>WP_044864167.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRGAADQRAETRTQAPGAVSQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGRPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSNSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFMALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKTGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPNFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVREIAGQERDRAAITEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRGAADQRAETRTQAPGAVSQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGRPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSNSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFMALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKTGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLLQRSGETPNFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVREIAGQERDRAAITEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 10249..37116
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| APS45_RS23475 (APS45_23680) | 10249..12465 | - | 2217 | WP_044864168 | type IV conjugative transfer system coupling protein TraD | virb4 |
| APS45_RS23480 (APS45_23685) | 12717..13448 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| APS45_RS23485 (APS45_23690) | 13462..13971 | - | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| APS45_RS23490 (APS45_23695) | 13968..16793 | - | 2826 | WP_001554363 | conjugal transfer mating-pair stabilization protein TraG | traG |
| APS45_RS23495 (APS45_23700) | 16790..18166 | - | 1377 | WP_000982833 | conjugal transfer pilus assembly protein TraH | traH |
| APS45_RS23500 (APS45_23705) | 18163..18525 | - | 363 | WP_001443292 | P-type conjugative transfer protein TrbJ | - |
| APS45_RS23505 (APS45_23710) | 18455..19000 | - | 546 | WP_000052618 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| APS45_RS23510 (APS45_23715) | 18987..19271 | - | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| APS45_RS23515 (APS45_23720) | 19390..19737 | - | 348 | WP_001287895 | conjugal transfer protein TrbA | - |
| APS45_RS23520 (APS45_23725) | 19753..20496 | - | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| APS45_RS23525 (APS45_23730) | 20489..20746 | - | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| APS45_RS23530 (APS45_23735) | 20773..22581 | - | 1809 | WP_000821818 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| APS45_RS23535 (APS45_23740) | 22578..23174 | - | 597 | WP_234034274 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| APS45_RS23540 (APS45_23745) | 23244..23696 | - | 453 | WP_000069427 | hypothetical protein | - |
| APS45_RS23545 (APS45_23750) | 23726..24187 | - | 462 | WP_000277845 | hypothetical protein | - |
| APS45_RS23550 (APS45_23755) | 24213..25205 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| APS45_RS23555 (APS45_23760) | 25202..25834 | - | 633 | WP_060554329 | type-F conjugative transfer system protein TraW | traW |
| APS45_RS23560 (APS45_23765) | 25831..26217 | - | 387 | WP_000214096 | type-F conjugative transfer system protein TrbI | - |
| APS45_RS23565 (APS45_23770) | 26214..28841 | - | 2628 | WP_001064248 | type IV secretion system protein TraC | virb4 |
| APS45_RS23570 (APS45_23775) | 29001..29222 | - | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| APS45_RS23575 (APS45_23780) | 29356..29871 | - | 516 | WP_000809906 | type IV conjugative transfer system lipoprotein TraV | traV |
| APS45_RS23580 (APS45_23785) | 29868..30119 | - | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| APS45_RS23585 (APS45_23790) | 30116..30433 | - | 318 | WP_001057292 | conjugal transfer protein TrbD | virb4 |
| APS45_RS23590 (APS45_23795) | 30430..31002 | - | 573 | WP_000002794 | conjugal transfer pilus-stabilizing protein TraP | - |
| APS45_RS23595 (APS45_23800) | 30992..32419 | - | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| APS45_RS23600 (APS45_23805) | 32419..33147 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| APS45_RS23605 (APS45_23810) | 33134..33700 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| APS45_RS23610 (APS45_23815) | 33722..34033 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| APS45_RS23615 (APS45_23820) | 34048..34407 | - | 360 | WP_000994781 | type IV conjugative transfer system pilin TraA | - |
| APS45_RS23620 (APS45_23825) | 34440..34835 | - | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| APS45_RS23625 (APS45_23830) | 34934..35623 | - | 690 | WP_000283380 | conjugal transfer transcriptional regulator TraJ | - |
| APS45_RS23630 (APS45_23835) | 35810..36193 | - | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| APS45_RS23635 (APS45_23840) | 36514..37116 | + | 603 | WP_000243703 | transglycosylase SLT domain-containing protein | virB1 |
| APS45_RS23640 (APS45_23845) | 37411..38232 | - | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| APS45_RS23645 (APS45_23850) | 38352..38639 | - | 288 | WP_000107535 | hypothetical protein | - |
| APS45_RS27005 (APS45_23855) | 38664..38870 | - | 207 | WP_000547971 | hypothetical protein | - |
| APS45_RS27340 | 38940..39113 | + | 174 | Protein_46 | hypothetical protein | - |
| APS45_RS27345 | 39111..39341 | - | 231 | WP_001426396 | hypothetical protein | - |
| APS45_RS23660 (APS45_23865) | 39561..39686 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| APS45_RS27350 | 39628..39777 | - | 150 | Protein_49 | plasmid maintenance protein Mok | - |
| APS45_RS23665 (APS45_23870) | 39956..40718 | - | 763 | Protein_50 | plasmid SOS inhibition protein A | - |
| APS45_RS23670 (APS45_23875) | 40715..41149 | - | 435 | WP_000845873 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 1205 | GenBank | NZ_CP012928 |
| Plasmid name | p12-4374_62 | Incompatibility group | IncFII |
| Plasmid size | 62920 bp | Coordinate of oriT [Strand] | 36265..36554 [+] |
| Host baterium | Salmonella enterica subsp. enterica serovar Heidelberg strain 12-4374 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |