Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100742 |
| Name | oriT_pCFSAN001588_002 |
| Organism | Salmonella enterica subsp. enterica serovar Cerro str. CFSAN001588 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012835 (47644..47753 [-], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GTAATTGTAATAGCGTC..GACGGTATTACAATTAC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pCFSAN001588_002
TCACTTCAGGCTCCTTACAGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACAGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file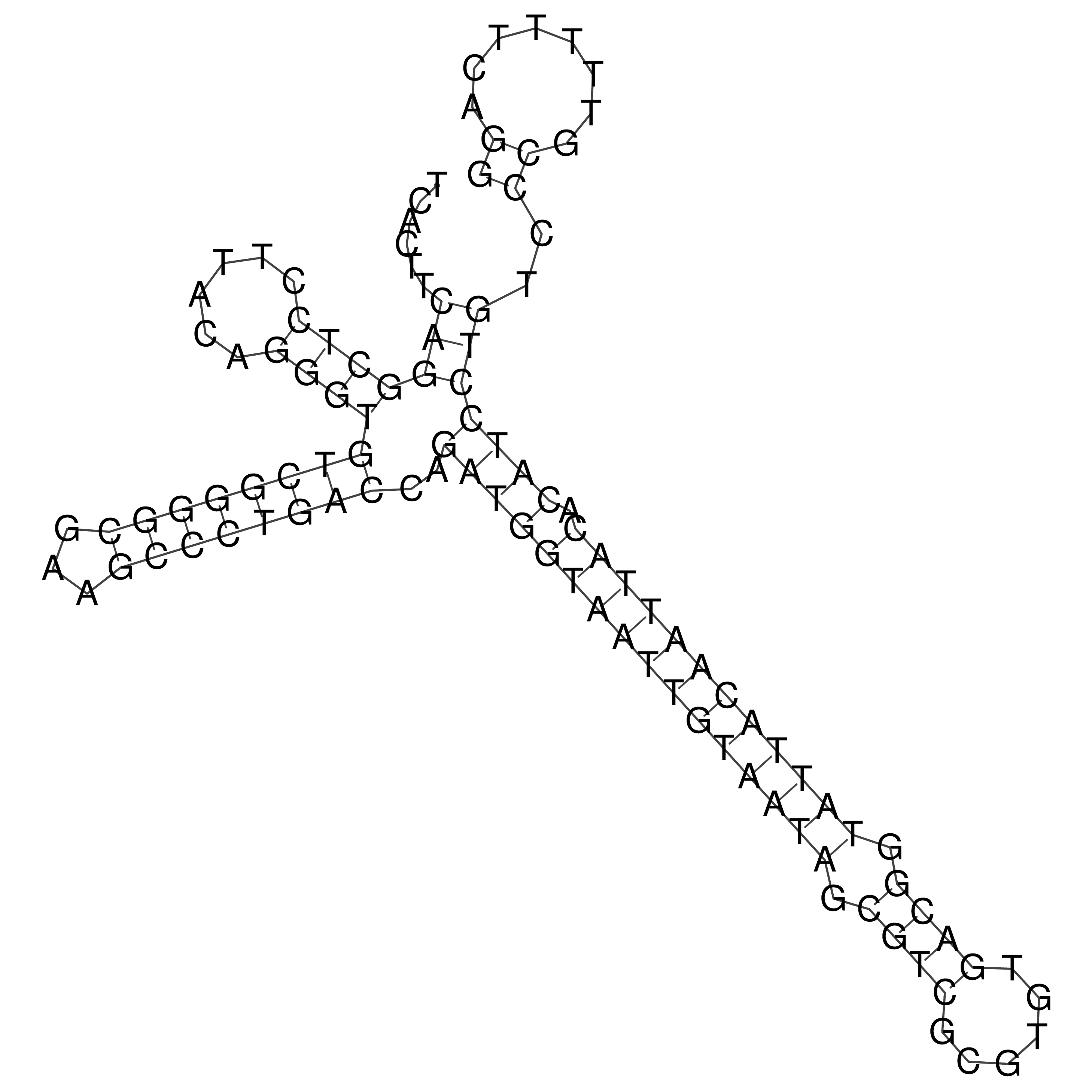
Relaxase
| ID | 814 | GenBank | WP_023233611 |
| Name | Nuclease_pCFSAN001588_002 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103945.58 Da Isoelectric Point: 7.4287
>WP_023233611.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRSEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVDAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLNGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYSDYIAANSNDEITLPSLELEKQWEPPKPM
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRSEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVDAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLNGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYSDYIAANSNDEITLPSLELEKQWEPPKPM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 1202 | GenBank | NZ_CP012835 |
| Plasmid name | pCFSAN001588_002 | Incompatibility group | IncI1 |
| Plasmid size | 62884 bp | Coordinate of oriT [Strand] | 47644..47753 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Cerro str. CFSAN001588 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |