Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100738 |
| Name | oriT_pSF-468-2 |
| Organism | Escherichia coli strain SF-468 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012627 (66217..66319 [+], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | IR1: 19..26, 30..37 (GTCGGGGC..GCCCTGAC) IR2: 44..58, 67..81 (GCAATTGTATAATCG..CGGTTATACAATTGC) |
| Location of nic site | 89..90 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 103 nt
>oriT_pSF-468-2
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
TCAGCTCCTTACTGGGGTGTCGGGGCGAAGCCCTGACCAGGAGGCAATTGTATAATCGCGCGTGCGCGGTTATACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file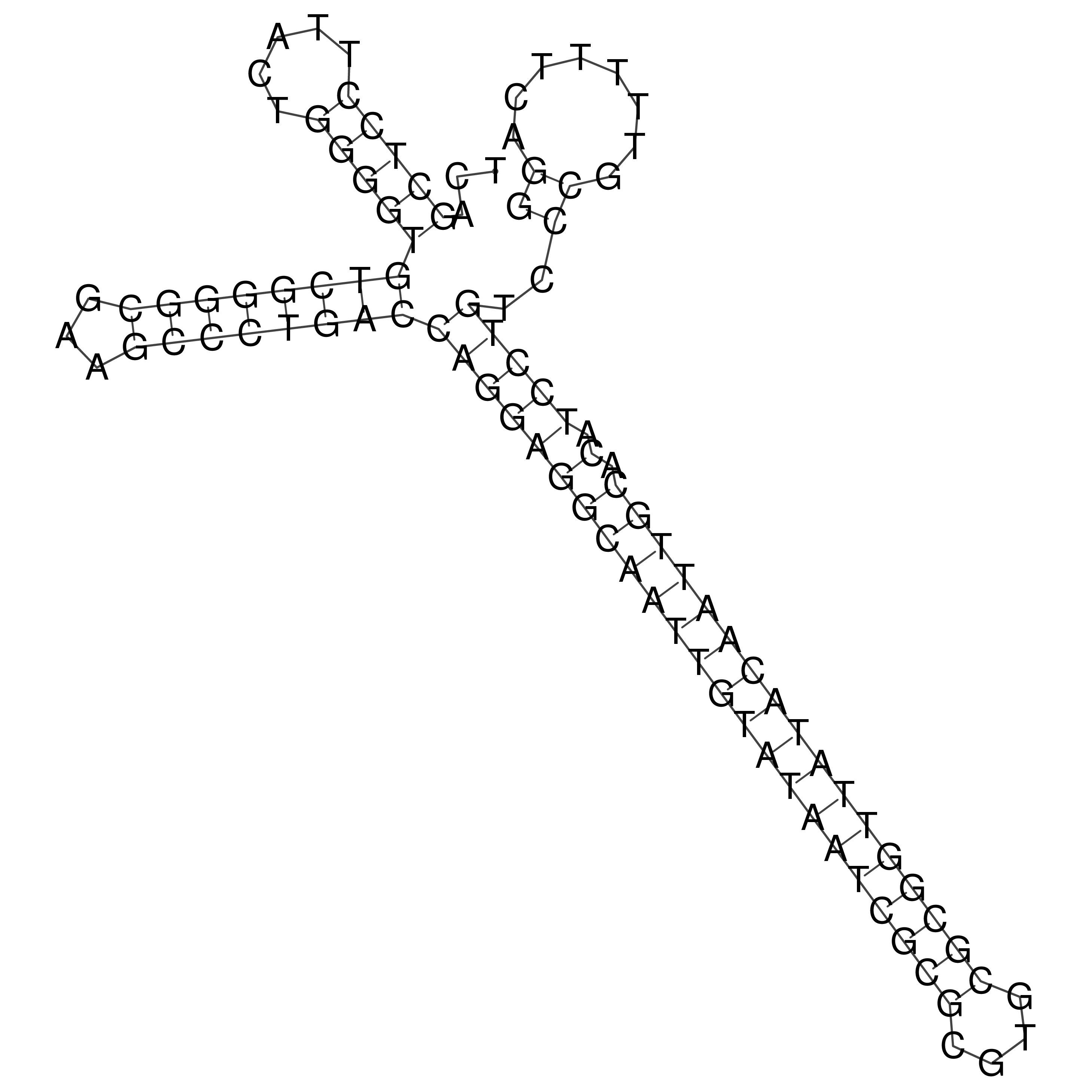
Relaxase
| ID | 810 | GenBank | WP_021520371 |
| Name | PutativerelaxaseofpSF-468-2 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103939.37 Da Isoelectric Point: 7.3426
>WP_021520371.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRNERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVNDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRNERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21181..60851
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AN206_RS26760 (AN206_26855) | 16243..17310 | + | 1068 | WP_012783947 | type IV pilus biogenesis lipoprotein PilL | - |
| AN206_RS26765 (AN206_26860) | 17310..17747 | + | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| AN206_RS26770 (AN206_26865) | 17761..19443 | + | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| AN206_RS26775 (AN206_26870) | 19436..20731 | + | 1296 | WP_001388364 | type 4b pilus protein PilO2 | - |
| AN206_RS26780 (AN206_26875) | 20718..21170 | + | 453 | WP_001247870 | type IV pilus biogenesis protein PilP | - |
| AN206_RS26785 (AN206_26880) | 21181..22734 | + | 1554 | WP_001492932 | ATPase, T2SS/T4P/T4SS family | virB11 |
| AN206_RS26790 (AN206_26885) | 22747..23844 | + | 1098 | WP_001492931 | type II secretion system F family protein | - |
| AN206_RS26795 (AN206_26890) | 23862..24476 | + | 615 | WP_001542527 | type 4 pilus major pilin | - |
| AN206_RS26800 (AN206_26895) | 24486..25046 | + | 561 | WP_000014116 | lytic transglycosylase domain-containing protein | virB1 |
| AN206_RS26805 (AN206_26900) | 25031..25687 | + | 657 | WP_001193553 | prepilin peptidase | - |
| AN206_RS26810 (AN206_26905) | 25687..26967 | + | 1281 | Protein_27 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| AN206_RS26815 (AN206_26910) | 28634..29788 | + | 1155 | WP_001139955 | site-specific integrase | - |
| AN206_RS26820 (AN206_26915) | 29939..30763 | + | 825 | WP_001238932 | conjugal transfer protein TraE | traE |
| AN206_RS26825 (AN206_26920) | 30849..32051 | + | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| AN206_RS26830 (AN206_26925) | 32111..32695 | + | 585 | WP_000977522 | histidine phosphatase family protein | - |
| AN206_RS26835 (AN206_26930) | 33089..33547 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| AN206_RS26840 (AN206_26935) | 33544..34362 | + | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| AN206_RS26845 (AN206_26940) | 34359..35507 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| AN206_RS29310 | 35504..35794 | + | 291 | WP_001299214 | hypothetical protein | traK |
| AN206_RS26850 (AN206_26945) | 35809..36360 | + | 552 | WP_000014583 | phospholipase D family protein | - |
| AN206_RS26855 (AN206_26950) | 36450..40217 | + | 3768 | WP_001141527 | LPD7 domain-containing protein | - |
| AN206_RS26860 (AN206_26955) | 40235..40582 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| AN206_RS26865 (AN206_26960) | 40579..41271 | + | 693 | WP_000138550 | DotI/IcmL family type IV secretion protein | traM |
| AN206_RS26870 (AN206_26965) | 41282..42265 | + | 984 | WP_001191877 | IncI1-type conjugal transfer protein TraN | traN |
| AN206_RS26875 (AN206_26970) | 42268..43557 | + | 1290 | WP_042004726 | conjugal transfer protein TraO | traO |
| AN206_RS26880 (AN206_26975) | 43557..44261 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| AN206_RS26885 (AN206_26980) | 44261..44788 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| AN206_RS26890 (AN206_26985) | 44839..45243 | + | 405 | WP_000086965 | IncI1-type conjugal transfer protein TraR | traR |
| AN206_RS26895 (AN206_26990) | 45307..45495 | + | 189 | WP_001277253 | putative conjugal transfer protein TraS | - |
| AN206_RS26900 (AN206_26995) | 45479..46279 | + | 801 | WP_001164784 | IncI1-type conjugal transfer protein TraT | traT |
| AN206_RS26905 (AN206_27000) | 46369..49413 | + | 3045 | WP_001024789 | IncI1-type conjugal transfer protein TraU | traU |
| AN206_RS26910 (AN206_27005) | 49413..50027 | + | 615 | WP_000337399 | IncI1-type conjugal transfer protein TraV | traV |
| AN206_RS26915 (AN206_27010) | 49994..51196 | + | 1203 | WP_042004725 | IncI1-type conjugal transfer protein TraW | traW |
| AN206_RS26920 (AN206_27015) | 51225..51809 | + | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| AN206_RS26925 (AN206_27020) | 51906..54074 | + | 2169 | WP_000698357 | DotA/TraY family protein | traY |
| AN206_RS26930 (AN206_27025) | 54148..54798 | + | 651 | WP_001178506 | plasmid IncI1-type surface exclusion protein ExcA | - |
| AN206_RS26935 (AN206_27030) | 54870..55079 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| AN206_RS31080 (AN206_27040) | 55471..55647 | + | 177 | WP_001054898 | hypothetical protein | - |
| AN206_RS31650 | 55712..55807 | - | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| AN206_RS26950 (AN206_27045) | 56306..56557 | + | 252 | WP_001291964 | hypothetical protein | - |
| AN206_RS26955 (AN206_27050) | 56629..56781 | - | 153 | WP_001331364 | Hok/Gef family protein | - |
| AN206_RS26960 (AN206_27055) | 57350..58336 | + | 987 | WP_001257838 | hypothetical protein | - |
| AN206_RS26965 (AN206_27060) | 58554..59762 | + | 1209 | WP_001383960 | IncI1-type conjugal transfer protein TrbA | trbA |
| AN206_RS26970 (AN206_27065) | 59781..60851 | + | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| AN206_RS26975 (AN206_27070) | 60844..63135 | + | 2292 | WP_021520372 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 1198 | GenBank | NZ_CP012627 |
| Plasmid name | pSF-468-2 | Incompatibility group | IncI1 |
| Plasmid size | 92766 bp | Coordinate of oriT [Strand] | 66217..66319 [+] |
| Host baterium | Escherichia coli strain SF-468 |
Cargo genes
| Drug resistance gene | blaCTX-M-14 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |