Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100733 |
| Name | oriT_pCFSAN000679_01 |
| Organism | Salmonella enterica subsp. enterica serovar Choleraesuis str. ATCC 10708 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012345 (102856..103114 [-], 259 nt) |
| oriT length | 259 nt |
| IRs (inverted repeats) | 115..128, 134..147 (ATAGCGGTGCCGGC..GCCTGCACCGCTAT) |
| Location of nic site | 241..242 |
| Conserved sequence flanking the nic site |
GTGGGGTGT|GG |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 259 nt
>oriT_pCFSAN000679_01
GCAGCGCCCCCAGGGGTGTTACTTAAAAACAGGCACAGCGACGCTAGCAGCGCCCCTGATATAATGCAATGTTTTTTATAAAAATAGTCAGTACTACCCCTGTATAGCGTCGTCATAGCGGTGCCGGCGACGTGCCTGCACCGCTATTTAATACTTTAAGATAAATTAAATTACATTATTATGAACATAACTCACTGTTACTAAAAACAAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
GCAGCGCCCCCAGGGGTGTTACTTAAAAACAGGCACAGCGACGCTAGCAGCGCCCCTGATATAATGCAATGTTTTTTATAAAAATAGTCAGTACTACCCCTGTATAGCGTCGTCATAGCGGTGCCGGCGACGTGCCTGCACCGCTATTTAATACTTTAAGATAAATTAAATTACATTATTATGAACATAACTCACTGTTACTAAAAACAAATCAGCAAAATTTTAATTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file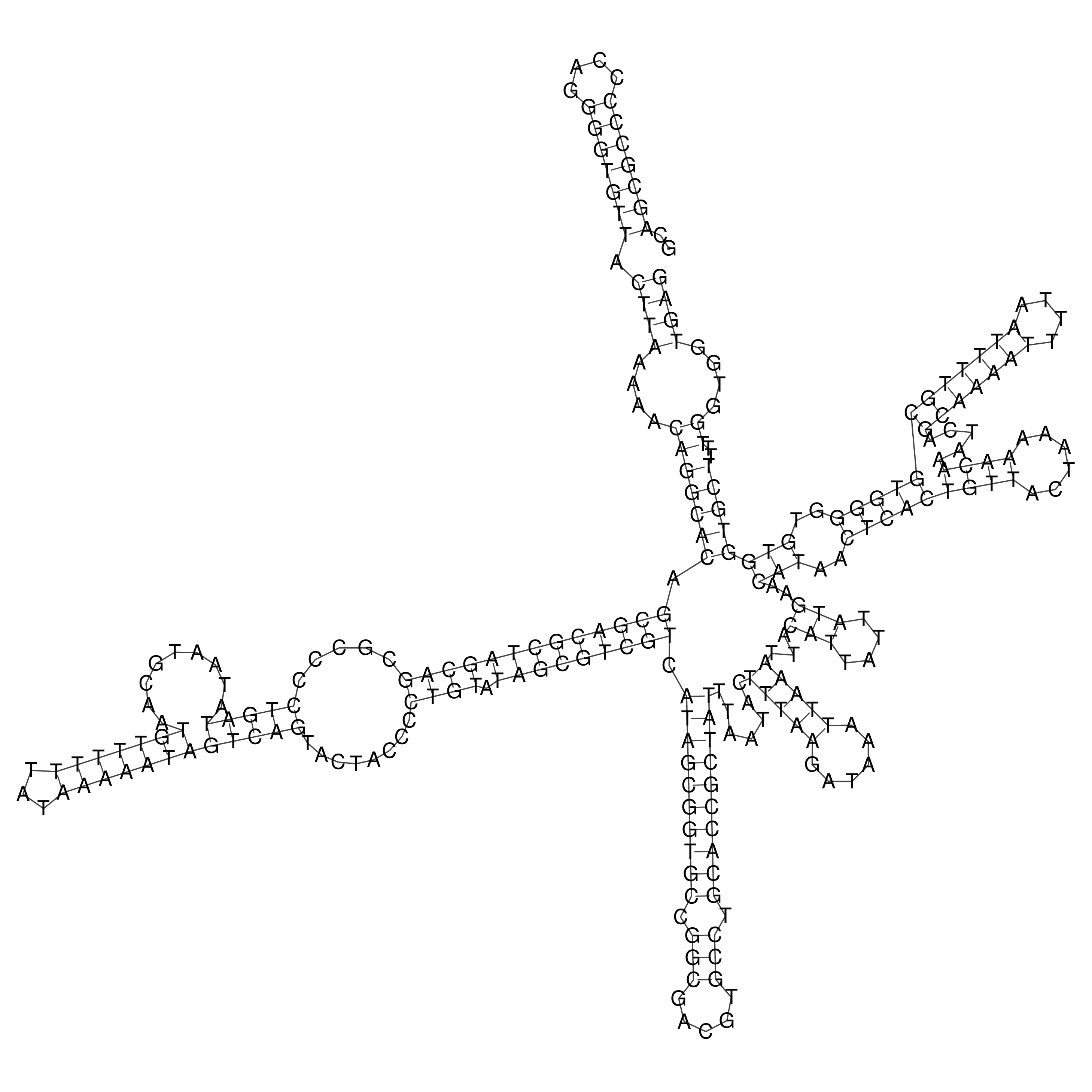
Relaxase
| ID | 805 | GenBank | WP_053253528 |
| Name | TraI_pCFSAN000679_01 |
UniProt ID | _ |
| Length | 1747 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1747 a.a. Molecular weight: 190612.51 Da Isoelectric Point: 5.9711
>WP_053253528.1 conjugative transfer relaxase/helicase TraI [Salmonella enterica]
MMSIAQVRSAGSAAGYYSDRDNYYVLGSLEERWAGKGAEQLGLQGAVDKEVFTRVLEGRLPDGADLSRQQ
DGSNKHRPGYDLTFSAPKSVSLMAMLAGDKRLTEAHNQAVDIAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTLHDGEWKTLSSDKVGKTGFIENVYANQIAFGKIYRAVLKEKVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQVIREAVGEDASLKSRDVAALDTRKSKQHVDPEVKMAEWMQT
LKDTGFDISAYREAADRRAEIQVAQPAPSQEQPDIQQAVTQAIAGLSDRKVQFTYTDVLARTVGMLSPEA
GVIEKARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSSDIMKQNRVTIHPEKSVPRTGSYSD
AVSVLAQDRPSLAIISGQGGAAGQRERVAELTMMAREQGREVQIIAADRRSQTNLKQDERLSGELITGRR
QLQEGMSFSPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKEAGVNSYRWQ
GGQQTPATVISEPDRNVRYARLAGDFVAAVKAGEESVAQVSGVREQAILAGMIRSELKTQGILGQQDTMM
TALSPVWLDSRNRYLRDMYREGMVMEQWNPEKRSHDRYVIDRVTAQSHSLTLRDAQGETQMVRISALDSS
WSLFRPEKIPVADGERLMVTGKIPGLRVSGGDRLQISAVNDGMMTVIVPGRAEPASLPVGDSPFTALKLE
NGWVETPGHSVSDSAKVFASVTQMAMNNATLNGLARSGRDVRLYSSLDETRTAEKLSRHPSFTVVSEQIK
ARAGEVSLETAISRQKAGLHTPAQQAIHLALPVVESKNLAFSQVELLTEAKSFAAVGTGFADLGREIDAQ
IKRGDLLHVDVAKGYGTDLLISRASYDAEKSILHHVLEGKEAVTPLMERVSGELMETLTSGQRAATRMIL
ETKDRFTVVQGYAGVGKTTQFRAVMSAVNLLPEDERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDTQ
LQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSAA
DVAIMKEIVRQTPELRDAVYSLINRDVNKALSGLENVKPIQVPRLKGAWVPENSVTEFSRLQERELVKAA
QEAEKKGEAFPDVPVTLYEAIVRDYTGRTPDAREQTLIVTHLNEDRRVLNSMTQDALAKPGEQQVTVPVL
TTANIRDGGLRRLSTWENHQGALALVDKVYHRIAGISKEDGLITLQDADGNTRLISPREAPAEGITLYNP
ETIRVGAGDRMRFTKSDRERGYVANSVWTVTAVSGDGVTLSDGKQTRVVRPGQDRAEQHIDLAYAITAHG
AQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWVKAINSAEQKGSAHDVLEPKS
EREMMNAERLFSTARELRDVAAGRAVLRNAGLAQGDSRARFIAPGRKYPQPYVALPAFDRNGKSAGIWLN
PLTTDDGAGLRGFSGEGRVKGSEEAQFVALQGSRNGESLLADNMQEGVRIARDNSDSGVVVRIAGDGRPW
NPGAITGGRVWGDIPDSSVQPGAGNGEPVTAEILAQRQAEEAVRRADEIVRKMAEDKSDLPEEKTAQAVR
EIAGQEQDRTLLPERKPPLPESVLREPVRERETILEVARESRVQERLQQMEQEMVRDLQKERTPGGD
MMSIAQVRSAGSAAGYYSDRDNYYVLGSLEERWAGKGAEQLGLQGAVDKEVFTRVLEGRLPDGADLSRQQ
DGSNKHRPGYDLTFSAPKSVSLMAMLAGDKRLTEAHNQAVDIAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTLHDGEWKTLSSDKVGKTGFIENVYANQIAFGKIYRAVLKEKVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQVIREAVGEDASLKSRDVAALDTRKSKQHVDPEVKMAEWMQT
LKDTGFDISAYREAADRRAEIQVAQPAPSQEQPDIQQAVTQAIAGLSDRKVQFTYTDVLARTVGMLSPEA
GVIEKARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSSDIMKQNRVTIHPEKSVPRTGSYSD
AVSVLAQDRPSLAIISGQGGAAGQRERVAELTMMAREQGREVQIIAADRRSQTNLKQDERLSGELITGRR
QLQEGMSFSPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKEAGVNSYRWQ
GGQQTPATVISEPDRNVRYARLAGDFVAAVKAGEESVAQVSGVREQAILAGMIRSELKTQGILGQQDTMM
TALSPVWLDSRNRYLRDMYREGMVMEQWNPEKRSHDRYVIDRVTAQSHSLTLRDAQGETQMVRISALDSS
WSLFRPEKIPVADGERLMVTGKIPGLRVSGGDRLQISAVNDGMMTVIVPGRAEPASLPVGDSPFTALKLE
NGWVETPGHSVSDSAKVFASVTQMAMNNATLNGLARSGRDVRLYSSLDETRTAEKLSRHPSFTVVSEQIK
ARAGEVSLETAISRQKAGLHTPAQQAIHLALPVVESKNLAFSQVELLTEAKSFAAVGTGFADLGREIDAQ
IKRGDLLHVDVAKGYGTDLLISRASYDAEKSILHHVLEGKEAVTPLMERVSGELMETLTSGQRAATRMIL
ETKDRFTVVQGYAGVGKTTQFRAVMSAVNLLPEDERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDTQ
LQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSAA
DVAIMKEIVRQTPELRDAVYSLINRDVNKALSGLENVKPIQVPRLKGAWVPENSVTEFSRLQERELVKAA
QEAEKKGEAFPDVPVTLYEAIVRDYTGRTPDAREQTLIVTHLNEDRRVLNSMTQDALAKPGEQQVTVPVL
TTANIRDGGLRRLSTWENHQGALALVDKVYHRIAGISKEDGLITLQDADGNTRLISPREAPAEGITLYNP
ETIRVGAGDRMRFTKSDRERGYVANSVWTVTAVSGDGVTLSDGKQTRVVRPGQDRAEQHIDLAYAITAHG
AQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWVKAINSAEQKGSAHDVLEPKS
EREMMNAERLFSTARELRDVAAGRAVLRNAGLAQGDSRARFIAPGRKYPQPYVALPAFDRNGKSAGIWLN
PLTTDDGAGLRGFSGEGRVKGSEEAQFVALQGSRNGESLLADNMQEGVRIARDNSDSGVVVRIAGDGRPW
NPGAITGGRVWGDIPDSSVQPGAGNGEPVTAEILAQRQAEEAVRRADEIVRKMAEDKSDLPEEKTAQAVR
EIAGQEQDRTLLPERKPPLPESVLREPVRERETILEVARESRVQERLQQMEQEMVRDLQKERTPGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 1193 | GenBank | NZ_CP012345 |
| Plasmid name | pCFSAN000679_01 | Incompatibility group | IncFIB |
| Plasmid size | 49584 bp | Coordinate of oriT [Strand] | 102856..103114 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Choleraesuis str. ATCC 10708 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |