Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100701 |
| Name | oriT_pAcX50c |
| Organism | Azotobacter chroococcum NCIMB 8003 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP010418 (12642..12731 [-], 90 nt) |
| oriT length | 90 nt |
| IRs (inverted repeats) | 1..19, 22..40 (GTGAAGATAGATAACCGGC..GCCGGTTAGCTAACTTCAC) |
| Location of nic site | 48..49 |
| Conserved sequence flanking the nic site |
TGTCTTG|C |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 90 nt
>oriT_pAcX50c
GTGAAGATAGATAACCGGCTTGCCGGTTAGCTAACTTCACCTGTCTTGCCCGGCTCTTCGAGCCGTTTAACGCCATATAAGTATCGCATA
GTGAAGATAGATAACCGGCTTGCCGGTTAGCTAACTTCACCTGTCTTGCCCGGCTCTTCGAGCCGTTTAACGCCATATAAGTATCGCATA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file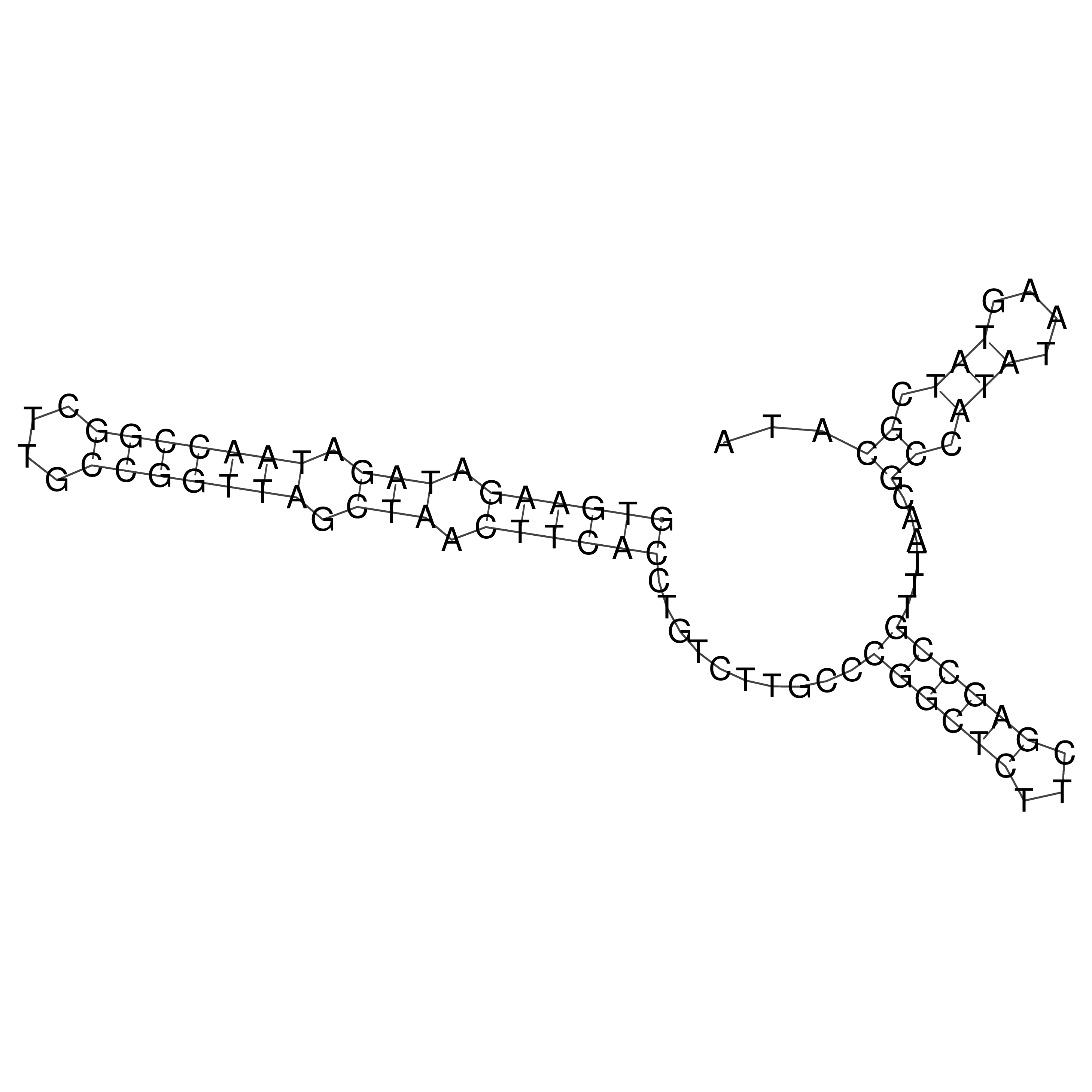
Relaxase
| ID | 773 | GenBank | WP_040107071 |
| Name | TraI_pAcX50c |
UniProt ID | A0A0C4WL02 |
| Length | 799 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 799 a.a. Molecular weight: 87713.68 Da Isoelectric Point: 10.8696
>WP_040107071.1 TraI/MobA(P) family conjugative relaxase [Azotobacter chroococcum]
MISKRIPMNSAKKSSFAGLVNYITNSQGISERVGEVRITNCQSESVTWAVRDILSAQQKNTRAEGDKTYH
LLISFAPGEKPSAEVLRDVEDRICAAMGYGEHQRISAVHNDTDVLHIHVAINKIHPQRLTMHEPYRDFKT
RAEICAKLEIEHGLERVNHTGRKSLSESKANDMERHAGVESLLSWVRRECLEQIQNATSWKEMHQVLHEH
GLELRKRGNGLVIADHEGTMVKASTVARDLSKNKLEDRLGEFEGRQHGRPTPSPGTPLPKPGIGKVGRRP
PPASRNRLRSMSSLQSLKIDSGKRYEKRPVGHRTDTTELYARYKDEQSAVRHTRAKELAAERDRRDRALE
NARRAADLKRASIKLMVAPGLNKKLLYASTHKTLKEQIAKIQTEHRKAVEKINTTRKARQWADWLQVKAK
EGDQEALKALRARAGVKGLQGDAIAAKGPQKPSQAVLAGQDHITKEGTVIYLAGAAAIRDDGSKLQVSRG
TTFDGIETALRMAVARYGKQVTVTGSPQFKELVAQTAAVRNLPIKFDDPALEQRRQALQQAIEKERNNVR
ADRGRAAGTSAGGNGFDPARRGRSDTGRTTGAADGQGDGVRGSATGTGVPERLGAVDGGRIPVRALGGTA
KPGVARVGRKPPPESQNRLRALSQLGVVQLTHGAEVLLPRHVPGDVEHQGAKPTDGLRRDVSRPGVAAPT
SPISGQQAARIYIEEREEKRAKGFDIPKHRLYNQADDGAASFEGVRQVKGQFMALLKRGDDVLVLPINEA
TARRMKRMKLGDPVRATAKGAIKSKGKSR
MISKRIPMNSAKKSSFAGLVNYITNSQGISERVGEVRITNCQSESVTWAVRDILSAQQKNTRAEGDKTYH
LLISFAPGEKPSAEVLRDVEDRICAAMGYGEHQRISAVHNDTDVLHIHVAINKIHPQRLTMHEPYRDFKT
RAEICAKLEIEHGLERVNHTGRKSLSESKANDMERHAGVESLLSWVRRECLEQIQNATSWKEMHQVLHEH
GLELRKRGNGLVIADHEGTMVKASTVARDLSKNKLEDRLGEFEGRQHGRPTPSPGTPLPKPGIGKVGRRP
PPASRNRLRSMSSLQSLKIDSGKRYEKRPVGHRTDTTELYARYKDEQSAVRHTRAKELAAERDRRDRALE
NARRAADLKRASIKLMVAPGLNKKLLYASTHKTLKEQIAKIQTEHRKAVEKINTTRKARQWADWLQVKAK
EGDQEALKALRARAGVKGLQGDAIAAKGPQKPSQAVLAGQDHITKEGTVIYLAGAAAIRDDGSKLQVSRG
TTFDGIETALRMAVARYGKQVTVTGSPQFKELVAQTAAVRNLPIKFDDPALEQRRQALQQAIEKERNNVR
ADRGRAAGTSAGGNGFDPARRGRSDTGRTTGAADGQGDGVRGSATGTGVPERLGAVDGGRIPVRALGGTA
KPGVARVGRKPPPESQNRLRALSQLGVVQLTHGAEVLLPRHVPGDVEHQGAKPTDGLRRDVSRPGVAAPT
SPISGQQAARIYIEEREEKRAKGFDIPKHRLYNQADDGAASFEGVRQVKGQFMALLKRGDDVLVLPINEA
TARRMKRMKLGDPVRATAKGAIKSKGKSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0C4WL02 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 54014..62621
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Achr_RS20875 (Achr_c420) | 49725..50747 | - | 1023 | WP_052264040 | RES family NAD+ phosphorylase | - |
| Achr_RS24535 (Achr_c430) | 51200..51814 | - | 615 | WP_082045612 | muramidase | - |
| Achr_RS24540 (Achr_c440) | 51818..52123 | - | 306 | WP_082045613 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| Achr_RS20880 (Achr_c450) | 52375..53752 | - | 1378 | Protein_46 | P-type conjugative transfer protein TrbL | - |
| Achr_RS20885 (Achr_c460) | 53762..54007 | - | 246 | WP_040107093 | entry exclusion lipoprotein TrbK | - |
| Achr_RS20890 (Achr_c470) | 54014..54799 | - | 786 | WP_040107094 | P-type conjugative transfer protein TrbJ | virB5 |
| Achr_RS20895 (Achr_c480) | 54823..56238 | - | 1416 | WP_040107095 | TrbI/VirB10 family protein | virB10 |
| Achr_RS20900 (Achr_c490) | 56245..56697 | - | 453 | WP_040107096 | conjugal transfer protein TrbH | - |
| Achr_RS20905 (Achr_c500) | 56701..57591 | - | 891 | WP_052264041 | P-type conjugative transfer protein TrbG | virB9 |
| Achr_RS20910 (Achr_c510) | 57603..58370 | - | 768 | WP_040107097 | conjugal transfer protein TrbF | virB8 |
| Achr_RS20915 (Achr_c520) | 58367..60925 | - | 2559 | WP_040107098 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| Achr_RS20920 (Achr_c530) | 60922..61233 | - | 312 | WP_040107099 | conjugal transfer protein TrbD | virB3 |
| Achr_RS20925 (Achr_c540) | 61236..61640 | - | 405 | WP_040107100 | conjugal transfer system pilin TrbC | virB2 |
| Achr_RS20930 (Achr_c550) | 61659..62621 | - | 963 | WP_040107101 | P-type conjugative transfer ATPase TrbB | virB11 |
Host bacterium
| ID | 1161 | GenBank | NZ_CP010418 |
| Plasmid name | pAcX50c | Incompatibility group | _ |
| Plasmid size | 62783 bp | Coordinate of oriT [Strand] | 12642..12731 [-] |
| Host baterium | Azotobacter chroococcum NCIMB 8003 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |