Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100689 |
| Name | oriT_pAPEC-O78-2 |
| Organism | Escherichia coli strain 789 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP010317 (32945..33054 [+], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GTAATTGTAATAGCGTC..GACGGTATTACAATTAC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pAPEC-O78-2
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file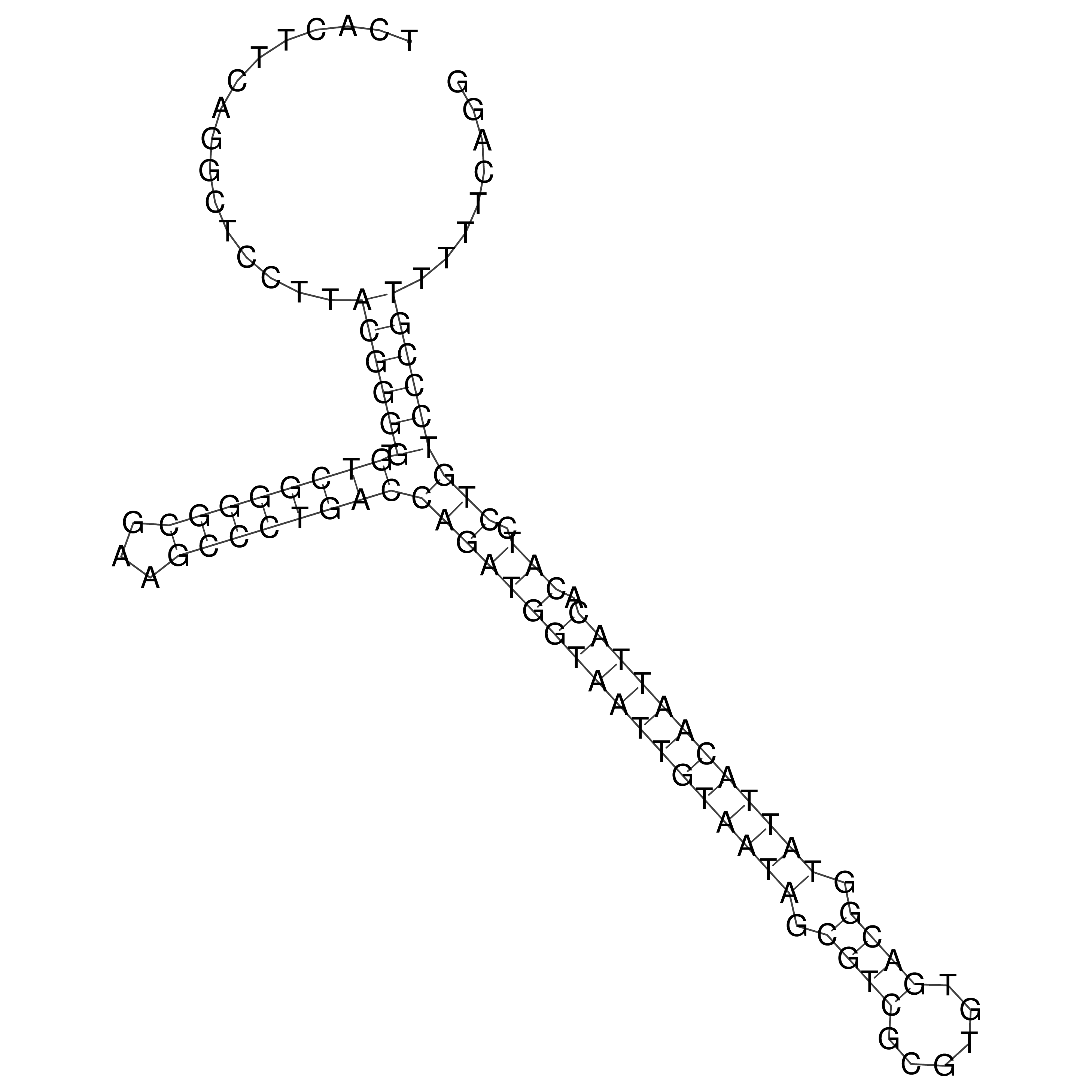
Relaxase
| ID | 761 | GenBank | WP_001474686 |
| Name | NikB_pAPEC-O78-2 |
UniProt ID | E9LLX8 |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 103834.33 Da Isoelectric Point: 7.3473
>WP_001474686.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFLQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | E9LLX8 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1485..27585
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AC789_RS25585 | 1..126 | + | 126 | WP_005119525 | IS1 family transposase | - |
| AC789_RS25590 (AC789_106pl00010) | 144..635 | + | 492 | Protein_1 | lipopolysaccharide core heptose(II)-phosphate phosphatase | - |
| AC789_RS25595 (AC789_106pl00020) | 1030..1488 | + | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| AC789_RS25600 (AC789_106pl00030) | 1485..2303 | + | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| AC789_RS25605 (AC789_106pl00040) | 2300..3448 | + | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| AC789_RS28385 | 3445..3735 | + | 291 | WP_001299214 | hypothetical protein | traK |
| AC789_RS25610 | 3750..4301 | + | 552 | WP_000014584 | phospholipase D family protein | - |
| AC789_RS25615 (AC789_106pl00060) | 4391..8158 | + | 3768 | WP_001141534 | LPD7 domain-containing protein | - |
| AC789_RS25620 (AC789_106pl00070) | 8176..8523 | + | 348 | WP_001055900 | conjugal transfer protein | traL |
| AC789_RS25625 (AC789_106pl00080) | 8520..9212 | + | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| AC789_RS25630 (AC789_106pl00090) | 9223..10206 | + | 984 | WP_001191878 | IncI1-type conjugal transfer protein TraN | traN |
| AC789_RS25635 (AC789_106pl00100) | 10209..11498 | + | 1290 | WP_001271997 | conjugal transfer protein TraO | traO |
| AC789_RS25640 (AC789_106pl00110) | 11498..12202 | + | 705 | WP_000801920 | IncI1-type conjugal transfer protein TraP | traP |
| AC789_RS25645 (AC789_106pl00120) | 12202..12729 | + | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| AC789_RS25650 (AC789_106pl00130) | 12780..13184 | + | 405 | WP_000086960 | IncI1-type conjugal transfer protein TraR | traR |
| AC789_RS25655 (AC789_106pl00140) | 13248..13436 | + | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| AC789_RS25660 (AC789_106pl00150) | 13420..14220 | + | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| AC789_RS25665 (AC789_106pl00160) | 14310..17354 | + | 3045 | WP_044502553 | IncI1-type conjugal transfer protein TraU | traU |
| AC789_RS25670 (AC789_106pl00170) | 17354..17968 | + | 615 | WP_000337400 | IncI1-type conjugal transfer protein TraV | traV |
| AC789_RS25675 (AC789_106pl00180) | 17935..19137 | + | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| AC789_RS25680 (AC789_106pl00190) | 19166..19750 | + | 585 | WP_001037985 | IncI1-type conjugal transfer protein TraX | - |
| AC789_RS25685 (AC789_106pl00200) | 19847..22015 | + | 2169 | WP_000698368 | DotA/TraY family protein | traY |
| AC789_RS25690 (AC789_106pl00210) | 22086..22748 | + | 663 | WP_000644794 | plasmid IncI1-type surface exclusion protein ExcA | - |
| AC789_RS25695 | 22820..23029 | - | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| AC789_RS30110 | 23421..23597 | + | 177 | WP_001054900 | hypothetical protein | - |
| AC789_RS25710 | 24521..24772 | + | 252 | WP_001291965 | hypothetical protein | - |
| AC789_RS25715 | 24844..24996 | - | 153 | WP_001303307 | Hok/Gef family protein | - |
| AC789_RS25720 (AC789_106pl00280) | 25288..26496 | + | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| AC789_RS25725 (AC789_106pl00290) | 26515..27585 | + | 1071 | WP_000151576 | IncI1-type conjugal transfer protein TrbB | trbB |
| AC789_RS25730 (AC789_106pl00300) | 27578..29869 | + | 2292 | WP_001289269 | F-type conjugative transfer protein TrbC | - |
Host bacterium
| ID | 1149 | GenBank | NZ_CP010317 |
| Plasmid name | pAPEC-O78-2 | Incompatibility group | IncI1 |
| Plasmid size | 106397 bp | Coordinate of oriT [Strand] | 32945..33054 [+] |
| Host baterium | Escherichia coli strain 789 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsH, arsA, arsC, arsB, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |