Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100670 |
| Name | oriT_pD3-B |
| Organism | Escherichia coli strain D3 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP010142 (26169..26278 [-], 110 nt) |
| oriT length | 110 nt |
| IRs (inverted repeats) | IR1: 24..31, 35..42 (GTCGGGGC..GCCCTGAC) IR2: 49..65, 72..88 (GCAATTGTAATAGCGTC..GACGGTATTACAATTGC) |
| Location of nic site | 96..97 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 110 nt
>oriT_pD3-B
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
TCACTTCAGGCTCCTTACGGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTCAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file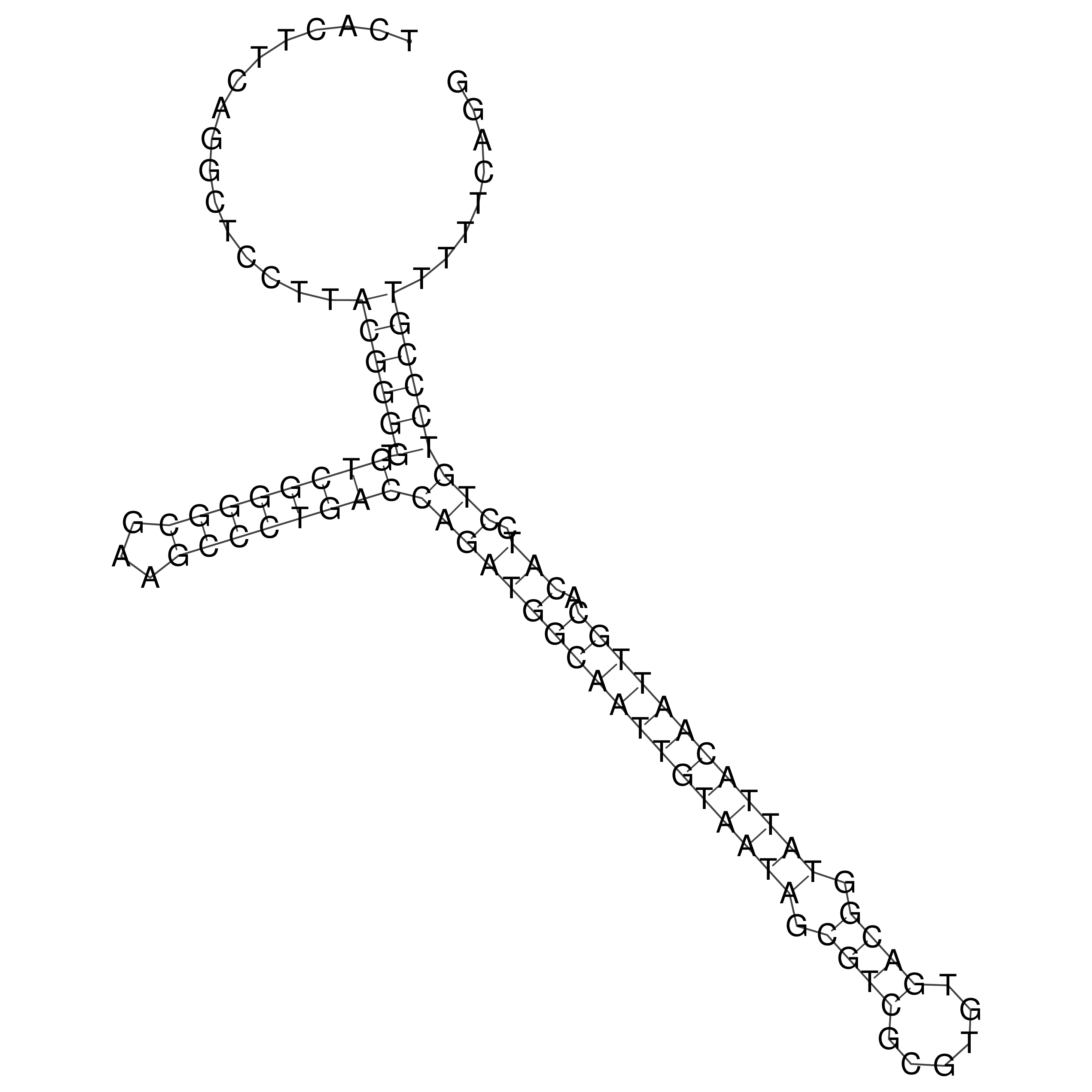
Relaxase
| ID | 742 | GenBank | WP_012783930 |
| Name | Relaxase_pD3-B |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104070.55 Da Isoelectric Point: 7.3623
>WP_012783930.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALVDKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALVDKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 31638..72547
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RG36_RS27630 (RG36_26040) | 29354..31645 | - | 2292 | WP_012783931 | F-type conjugative transfer protein TrbC | - |
| RG36_RS27635 (RG36_26045) | 31638..32708 | - | 1071 | WP_012783932 | IncI1-type conjugal transfer protein TrbB | trbB |
| RG36_RS27640 (RG36_26050) | 32727..33935 | - | 1209 | WP_001703845 | IncI1-type conjugal transfer protein TrbA | trbA |
| RG36_RS27645 (RG36_26055) | 34242..35021 | - | 780 | WP_275450201 | protein FinQ | - |
| RG36_RS27650 (RG36_26060) | 35648..35800 | + | 153 | WP_001387489 | Hok/Gef family protein | - |
| RG36_RS27655 (RG36_26065) | 35872..36123 | - | 252 | WP_001291964 | hypothetical protein | - |
| RG36_RS30080 | 36624..36719 | + | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| RG36_RS29550 (RG36_26075) | 36784..36960 | - | 177 | WP_001054900 | hypothetical protein | - |
| RG36_RS27675 (RG36_26085) | 37352..37561 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| RG36_RS27680 (RG36_26090) | 37633..38295 | - | 663 | WP_012783935 | plasmid IncI1-type surface exclusion protein ExcA | - |
| RG36_RS27685 (RG36_26095) | 38366..40534 | - | 2169 | WP_015508354 | DotA/TraY family protein | traY |
| RG36_RS27690 (RG36_26100) | 40631..41215 | - | 585 | WP_012783937 | conjugal transfer protein TraX | - |
| RG36_RS27695 (RG36_26105) | 41244..42446 | - | 1203 | WP_012783938 | IncI1-type conjugal transfer protein TraW | traW |
| RG36_RS27700 (RG36_26110) | 42413..43027 | - | 615 | WP_000337394 | IncI1-type conjugal transfer protein TraV | traV |
| RG36_RS27705 (RG36_26115) | 43027..46071 | - | 3045 | WP_012783939 | IncI1-type conjugal transfer protein TraU | traU |
| RG36_RS27710 (RG36_26120) | 46161..46961 | - | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| RG36_RS27715 (RG36_26125) | 46945..47133 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| RG36_RS27720 (RG36_26130) | 47197..47601 | - | 405 | WP_000086965 | IncI1-type conjugal transfer protein TraR | traR |
| RG36_RS27725 (RG36_26135) | 47652..48179 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| RG36_RS27730 (RG36_26140) | 48179..48883 | - | 705 | WP_001493811 | IncI1-type conjugal transfer protein TraP | traP |
| RG36_RS27735 (RG36_26145) | 48883..50172 | - | 1290 | WP_011264042 | conjugal transfer protein TraO | traO |
| RG36_RS27740 (RG36_26150) | 50175..51158 | - | 984 | WP_001191880 | IncI1-type conjugal transfer protein TraN | traN |
| RG36_RS27745 (RG36_26155) | 51169..51861 | - | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| RG36_RS27750 (RG36_26160) | 51858..52205 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| RG36_RS27755 (RG36_26165) | 52223..55990 | - | 3768 | WP_012783940 | LPD7 domain-containing protein | - |
| RG36_RS27760 (RG36_26170) | 56080..56301 | - | 222 | Protein_66 | phospholipase D-like domain-containing protein | - |
| RG36_RS27765 (RG36_26175) | 56388..56813 | + | 426 | WP_000422741 | transposase | - |
| RG36_RS27770 (RG36_26180) | 56810..57160 | + | 351 | WP_000624722 | IS66 family insertion sequence element accessory protein TnpB | - |
| RG36_RS27775 (RG36_26185) | 57191..58804 | + | 1614 | WP_000080200 | IS66-like element ISEc23 family transposase | - |
| RG36_RS27780 (RG36_26190) | 58836..59171 | - | 336 | Protein_70 | phospholipase D-like domain-containing protein | - |
| RG36_RS27785 | 59186..59476 | - | 291 | WP_001314267 | hypothetical protein | traK |
| RG36_RS27790 (RG36_26195) | 59473..60621 | - | 1149 | WP_001024971 | plasmid transfer ATPase TraJ | virB11 |
| RG36_RS27795 (RG36_26200) | 60618..61436 | - | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| RG36_RS27800 (RG36_26205) | 61433..61891 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| RG36_RS27805 (RG36_26210) | 62285..62869 | - | 585 | WP_000977522 | histidine phosphatase family protein | - |
| RG36_RS27810 (RG36_26215) | 62929..64131 | - | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| RG36_RS27815 (RG36_26220) | 64217..65041 | - | 825 | WP_001238932 | conjugal transfer protein TraE | traE |
| RG36_RS27820 (RG36_26225) | 65192..66346 | - | 1155 | WP_001139955 | site-specific integrase | - |
| RG36_RS29950 | 66345..66752 | + | 408 | WP_003552681 | hypothetical protein | - |
| RG36_RS27830 (RG36_26230) | 66749..68041 | - | 1293 | WP_001417545 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| RG36_RS27835 (RG36_26235) | 68041..68697 | - | 657 | WP_001193553 | prepilin peptidase | - |
| RG36_RS27840 (RG36_26240) | 68682..69242 | - | 561 | WP_000014116 | lytic transglycosylase domain-containing protein | virB1 |
| RG36_RS27845 (RG36_26245) | 69252..69866 | - | 615 | WP_001542527 | type 4 pilus major pilin | - |
| RG36_RS27850 (RG36_26250) | 69884..70981 | - | 1098 | WP_001492931 | type II secretion system F family protein | - |
| RG36_RS27855 (RG36_26255) | 70994..72547 | - | 1554 | WP_001492932 | ATPase, T2SS/T4P/T4SS family | virB11 |
| RG36_RS27860 (RG36_26260) | 72558..73010 | - | 453 | WP_012783945 | type IV pilus biogenesis protein PilP | - |
| RG36_RS27865 (RG36_26265) | 72997..74292 | - | 1296 | WP_012783946 | type 4b pilus protein PilO2 | - |
| RG36_RS27870 (RG36_26270) | 74285..75967 | - | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| RG36_RS27875 (RG36_26275) | 75981..76418 | - | 438 | WP_000539807 | type IV pilus biogenesis protein PilM | - |
| RG36_RS27880 (RG36_26280) | 76418..77485 | - | 1068 | WP_012783947 | type IV pilus biogenesis lipoprotein PilL | - |
Host bacterium
| ID | 1130 | GenBank | NZ_CP010142 |
| Plasmid name | pD3-B | Incompatibility group | IncI1 |
| Plasmid size | 90974 bp | Coordinate of oriT [Strand] | 26169..26278 [-] |
| Host baterium | Escherichia coli strain D3 |
Cargo genes
| Drug resistance gene | blaCTX-M-55, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrVA2 |