Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100641 |
| Name | oriT_pCTN1046 |
| Organism | Ligilactobacillus salivarius strain JCM1046 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP007650 (1792..2006 [+], 215 nt) |
| oriT length | 215 nt |
| IRs (inverted repeats) | 61..76, 80..95 (ACTTAACCCCCCGTAT..ACAGGGGGGTACAAAT) 123..134, 139..150 (GAAAATCCTTTG..CAAGGGATTTAC) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 215 nt
>oriT_pCTN1046
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file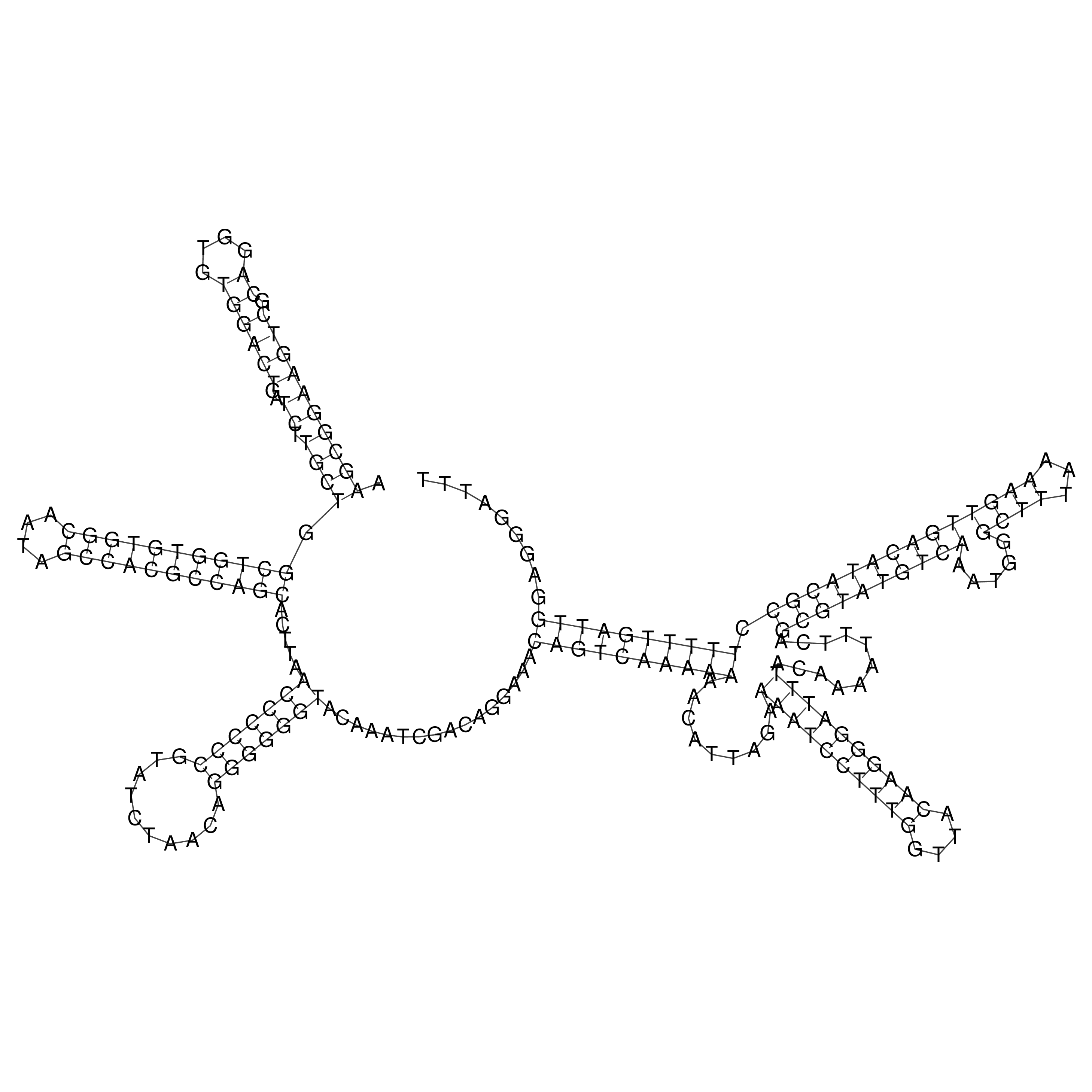
Relaxase
| ID | 713 | GenBank | WP_000398284 |
| Name | Relaxase_pCTN1046 |
UniProt ID | A0A089QLZ9 |
| Length | 401 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A089QLZ9 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 432..11097
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LSJ_RS12035 | 317..403 | + | 87 | Protein_0 | conjugal transfer protein | - |
| LSJ_RS11385 (LSJ_5001) | 432..1817 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| LSJ_RS11390 | 1820..1972 | + | 153 | WP_000879507 | hypothetical protein | - |
| LSJ_RS11395 (LSJ_5002) | 1995..3200 | + | 1206 | WP_000398284 | MobT family relaxase | - |
| LSJ_RS11400 (LSJ_5003) | 3243..3464 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| LSJ_RS11405 (LSJ_5004) | 3581..4078 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| LSJ_RS11410 (LSJ_5005) | 4167..4559 | + | 393 | WP_000723888 | conjugal transfer protein | orf17a |
| LSJ_RS11415 (LSJ_5006) | 4543..6990 | + | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| LSJ_RS11420 (LSJ_5007) | 6993..9170 | + | 2178 | WP_000804748 | membrane protein | orf15 |
| LSJ_RS11425 (LSJ_5008) | 9167..10168 | + | 1002 | WP_000769868 | bifunctional lysozyme/C40 family peptidase | orf14 |
| LSJ_RS11430 (LSJ_5009) | 10165..11097 | + | 933 | WP_001224318 | conjugal transfer protein | orf13 |
| LSJ_RS12040 (LSJ_5010) | 11342..11458 | + | 117 | WP_001791010 | tetracycline resistance determinant leader peptide | - |
| LSJ_RS11435 (LSJ_5011) | 11474..13393 | + | 1920 | WP_002371500 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| LSJ_RS12045 (LSJ_5012) | 13512..13679 | + | 168 | WP_000336323 | cysteine-rich KTR domain-containing protein | - |
| LSJ_RS11445 (LSJ_5013c) | 13739..14092 | - | 354 | WP_001227347 | helix-turn-helix transcriptional regulator | - |
| LSJ_RS11450 (LSJ_5015) | 14597..15019 | + | 423 | WP_000804885 | sigma-70 family RNA polymerase sigma factor | - |
| LSJ_RS11455 (LSJ_5016) | 15016..15246 | + | 231 | WP_000857133 | helix-turn-helix domain-containing protein | - |
| LSJ_RS12880 (LSJ_5018c) | 15472..15723 | - | 252 | WP_001845478 | hypothetical protein | - |
| LSJ_RS11460 (LSJ_5019) | 15707..15910 | + | 204 | WP_000814511 | excisionase | - |
Host bacterium
| ID | 1101 | GenBank | NZ_CP007650 |
| Plasmid name | pCTN1046 | Incompatibility group | _ |
| Plasmid size | 33315 bp | Coordinate of oriT [Strand] | 1792..2006 [+] |
| Host baterium | Ligilactobacillus salivarius strain JCM1046 |
Cargo genes
| Drug resistance gene | tetracycline resistance |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |