Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100630 |
| Name | oriT_pCC1557 |
| Organism | Pseudomonas syringae CC1557 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP007015 (26868..27013 [+], 146 nt) |
| oriT length | 146 nt |
| IRs (inverted repeats) | IR1: 38..43, 52..57 (AAAAAG..CTTTTT) IR2: 72..83, 87..98 (GTGCCAAAGGGG..CCCCTTTGAAAC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 146 nt
>oriT_pCC1557
CGAAATCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCGGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCAT
CGAAATCGCTAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTAGCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCGGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file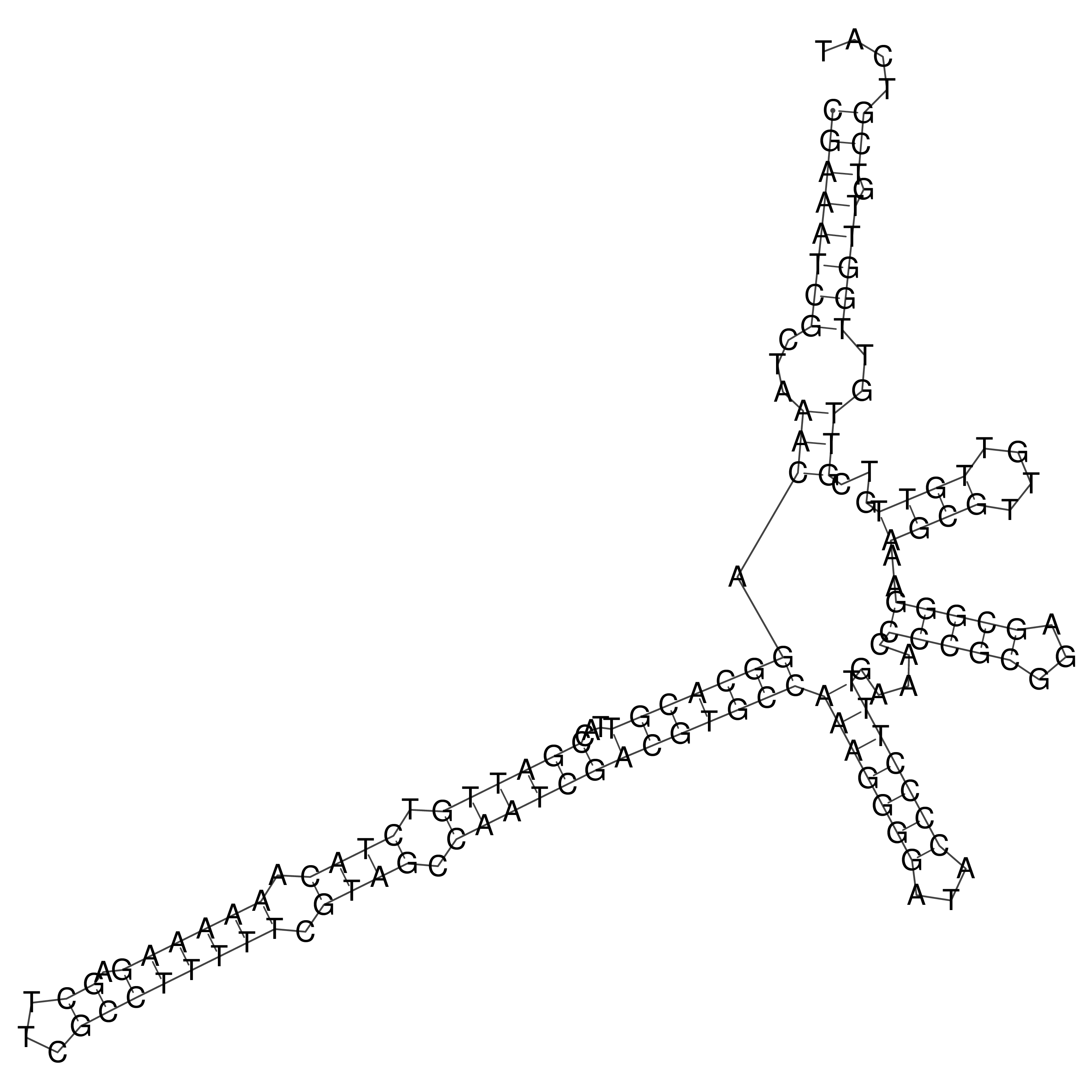
Relaxase
| ID | 702 | GenBank | WP_080274973 |
| Name | Relaxase_pCC1557 |
UniProt ID | _ |
| Length | 1249 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1249 a.a. Molecular weight: 138427.86 Da Isoelectric Point: 9.3676
>WP_080274973.1 LPD7 domain-containing protein [Pseudomonas syringae]
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDAFIVRRELKKPPLEASVIQD
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERENNFYRAHGDDYESVHTWQRQRDNQRSPA
ETAGRRTAAPADGLQDLSVSDVADHRQAGPTRSRDGALLLPSDAHVHVGQSQPGGDSGLRSSVPAGGRGR
RAGSTTERGRGGSQPAAVSQETKGTATPAGTTGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSRPQVAGQLPQARAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKALVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPAPEGVRERVKRVMDRFAKQVDPVAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADHLGVTVPIEALYGQGKTADQVSQALATQL
DTVPEPERIAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAANSAASATSEAQANVPTPSDAVSEPV
SPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEAEVRASDTVMGLRGEDHAVWIIATNDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPKLVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEEAMSDADLKQGDQVRLQDLGTQPVVV
QVIEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAAAAGTPELSSPDEDDGMSVD
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDRKTGEVIDRKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQQLVKQVIER
GVTSRADFYALVAEHGETRIRNEGKDNEYISVKLPGDAKGTNLKDTIFQDAFIVRRELKKPPLEASVIQD
RLLAWPQRAREIKYVNKATPKFRKQYSQASPEERVRLLAERENNFYRAHGDDYESVHTWQRQRDNQRSPA
ETAGRRTAAPADGLQDLSVSDVADHRQAGPTRSRDGALLLPSDAHVHVGQSQPGGDSGLRSSVPAGGRGR
RAGSTTERGRGGSQPAAVSQETKGTATPAGTTGRRRAGAGKPRNTRVRAGRVVPPYAQNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRSSSRPQVAGQLPQARAATTTPVQSTAGRRPRSSGKPRPPRQWRPGAVP
PYAKNPHRVATVADIEQRARMLFDPLKRPADKALVFKRASIKALTVNKHASTVAAYFTRQAQHNQIAPAH
RRAIRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDD
EKEDPGFSISGARGPAPEGVRERVKRVMDRFAKQVDPVAASERARDLSAKDLYTRKAKFSQNVHYLDKQT
DKTLFVDTGTTISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMN
QSLADRRAELDIERDGQSIGPATDLPRHVDDATRDVRDQADHLGVTVPIEALYGQGKTADQVSQALATQL
DTVPEPERIAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAANSAASATSEAQANVPTPSDAVSEPV
SPEAAKPALANDPDLQSPSELVRLEAQWRRDFPMSEAEVRASDTVMGLRGEDHAVWIIATNDKTPEAAAL
LTAYMENDSYREAFKASIVAAYKQVENSPKLVDDLDHLTAMAAQIVNEVEERLFPTPQAATGQTAPSRSK
VIEGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEEAMSDADLKQGDQVRLQDLGTQPVVV
QVIEEDGTVTDKTVNRREWSAQPVAPEREVAETTPKGQAAAAGTPELSSPDEDDGMSVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5880..16246
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| N018_RS25380 (N018_25870) | 1208..1534 | + | 327 | WP_025391071 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| N018_RS25385 (N018_25875) | 1646..1933 | + | 288 | WP_025391072 | XRE family transcriptional regulator | - |
| N018_RS25390 (N018_25880) | 2123..2896 | - | 774 | WP_017279416 | helix-turn-helix transcriptional regulator | - |
| N018_RS25395 (N018_25885) | 3005..4105 | + | 1101 | WP_025391073 | polyamine ABC transporter substrate-binding protein | - |
| N018_RS25400 (N018_25890) | 4202..4399 | + | 198 | WP_025391074 | hypothetical protein | - |
| N018_RS25405 (N018_25895) | 4411..4776 | - | 366 | WP_025391075 | helix-turn-helix transcriptional regulator | - |
| N018_RS25410 (N018_25900) | 5024..5536 | + | 513 | WP_010199861 | transcription termination/antitermination NusG family protein | - |
| N018_RS25415 (N018_25905) | 5625..5867 | + | 243 | WP_017279420 | hypothetical protein | - |
| N018_RS25420 (N018_25910) | 5880..6587 | + | 708 | WP_025391076 | lytic transglycosylase domain-containing protein | virB1 |
| N018_RS25425 (N018_25915) | 6587..6922 | + | 336 | WP_025391077 | hypothetical protein | - |
| N018_RS25430 (N018_25920) | 6935..7411 | + | 477 | WP_040118647 | VirB3 family type IV secretion system protein | virB3 |
| N018_RS25435 (N018_25925) | 7326..9824 | + | 2499 | WP_229631655 | conjugal transfer protein | virb4 |
| N018_RS25440 (N018_25930) | 9821..10507 | + | 687 | WP_025391079 | P-type DNA transfer protein VirB5 | - |
| N018_RS25445 (N018_25935) | 10536..10889 | + | 354 | WP_025391080 | hypothetical protein | - |
| N018_RS25450 (N018_25940) | 10900..11835 | + | 936 | WP_025391081 | type IV secretion system protein | virB6 |
| N018_RS25455 (N018_25945) | 12261..13049 | + | 789 | WP_025391082 | type IV secretion system protein | virB8 |
| N018_RS25460 (N018_25950) | 13039..13848 | + | 810 | WP_025391083 | P-type conjugative transfer protein VirB9 | virB9 |
| N018_RS25465 (N018_25955) | 13835..15205 | + | 1371 | WP_025391084 | TrbI/VirB10 family protein | virB10 |
| N018_RS25470 (N018_25960) | 15215..16246 | + | 1032 | WP_025391085 | P-type DNA transfer ATPase VirB11 | virB11 |
| N018_RS25475 (N018_25965) | 16256..16612 | + | 357 | WP_010217013 | hypothetical protein | - |
| N018_RS25480 (N018_25970) | 16738..19128 | + | 2391 | WP_025391086 | ATP-binding protein | - |
| N018_RS25485 (N018_25975) | 19215..19445 | + | 231 | WP_025391087 | hypothetical protein | - |
| N018_RS25490 (N018_25980) | 19445..21097 | + | 1653 | WP_025391088 | type IV secretory system conjugative DNA transfer family protein | - |
Host bacterium
| ID | 1090 | GenBank | NZ_CP007015 |
| Plasmid name | pCC1557 | Incompatibility group | _ |
| Plasmid size | 53629 bp | Coordinate of oriT [Strand] | 26868..27013 [+] |
| Host baterium | Pseudomonas syringae CC1557 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |