Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100613 |
| Name | oriT_p92-9000 |
| Organism | Shigella dysenteriae |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_032099 (24104..24189 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | IR1: 7..15, 17..25 (GTCGGGGCT..AGCCCTGAC) IR2: 32..49, 52..69 (GTAATCGTATAGCCGTGC..GCGCGGTTATACGATTAC) |
| Location of nic site | 77..78 |
| Conserved sequence flanking the nic site |
CATCCTG|T |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 86 nt
>oriT_p92-9000
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
CGGGGTGTCGGGGCTAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCGTTTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file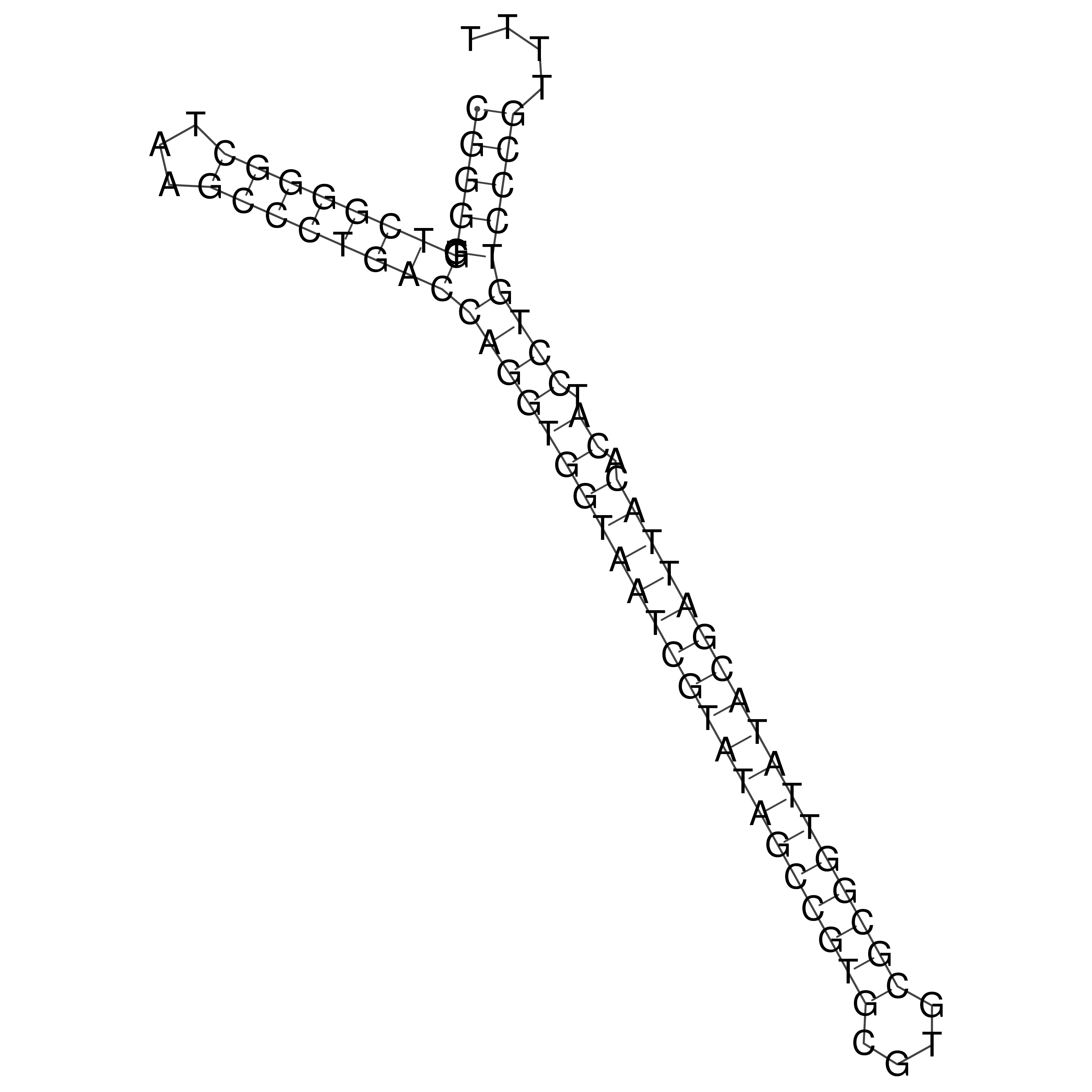
Relaxase
| ID | 685 | GenBank | YP_009328414 |
| Name | Putativerelaxaseofp92-9000 |
UniProt ID | A0A142CND5 |
| Length | 906 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 104908.50 Da Isoelectric Point: 8.2690
>YP_009328414.1 hypothetical protein (plasmid) [Shigella dysenteriae 1]
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVEAPGRYDPEAINVDIRPEKVAETE
SLKQYASRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPKKRRDGKSSFEDLIAYTSVRDDVPEDELRPDTEVKGEVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCLSLESAAEEMQKAADLAHFSKDDTDPVFHYILSWPAHESPRSEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINCLPWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGIALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVEAPGRYDPEAINVDIRPEKVAETE
SLKQYASRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVVPGERFRGHSIGSQPVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
GLIQCRSLIEEMIADGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPEIRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAYIRVQFRDPQLRKLHYHIAEVQRMQALIRLKESVKEERLSLIAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNVDRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLNLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CND5 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 29770..68452
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HPG17_RS00170 (BTU53_p045) | 27489..29789 | - | 2301 | WP_000077653 | F-type conjugative transfer protein TrbC | - |
| HPG17_RS00175 (BTU53_p046) | 29770..30894 | - | 1125 | WP_000152384 | DsbC family protein | trbB |
| HPG17_RS00180 (BTU53_p047) | 30891..32216 | - | 1326 | WP_171265269 | IncI1-type conjugal transfer protein TrbA | trbA |
| HPG17_RS00680 | 32753..32848 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HPG17_RS00685 | 33437..33532 | + | 96 | WP_001363141 | DinQ-like type I toxin DqlB | - |
| HPG17_RS00190 (BTU53_p052) | 33595..33771 | - | 177 | WP_001054907 | hypothetical protein | - |
| HPG17_RS00195 (BTU53_p053) | 33922..34389 | - | 468 | WP_000598962 | thermonuclease family protein | - |
| HPG17_RS00200 (BTU53_p054) | 34549..35172 | + | 624 | WP_001155250 | helix-turn-helix domain-containing protein | - |
| HPG17_RS00205 (BTU53_p055) | 35370..35585 | - | 216 | WP_000266543 | hypothetical protein | - |
| HPG17_RS00210 (BTU53_p056) | 35589..35957 | - | 369 | WP_001363143 | hypothetical protein | - |
| HPG17_RS00215 (BTU53_p058) | 36257..36499 | - | 243 | WP_001363144 | hypothetical protein | - |
| HPG17_RS00220 (BTU53_p060) | 36829..36981 | + | 153 | WP_001302699 | Hok/Gef family protein | - |
| HPG17_RS00225 (BTU53_p063) | 37608..38255 | - | 648 | WP_015056434 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HPG17_RS00230 (BTU53_p064) | 38343..40502 | - | 2160 | WP_000691811 | DotA/TraY family protein | traY |
| HPG17_RS00235 (BTU53_p065) | 40577..41146 | - | 570 | WP_000650752 | conjugal transfer protein TraX | - |
| HPG17_RS00240 (BTU53_p066) | 41143..42348 | - | 1206 | WP_001189548 | conjugal transfer protein TraW | traW |
| HPG17_RS00245 (BTU53_p067) | 42306..42926 | - | 621 | WP_048967127 | hypothetical protein | traV |
| HPG17_RS00250 (BTU53_p068) | 42926..45970 | - | 3045 | WP_001024766 | conjugal transfer protein | traU |
| HPG17_RS00255 (BTU53_p070) | 46265..47083 | - | 819 | WP_000753550 | hypothetical protein | traT |
| HPG17_RS00675 | 46998..47282 | - | 285 | WP_001353683 | hypothetical protein | - |
| HPG17_RS00265 (BTU53_p072) | 47306..47704 | - | 399 | WP_000088813 | DUF6750 family protein | traR |
| HPG17_RS00270 (BTU53_p073) | 47751..48281 | - | 531 | WP_000987021 | conjugal transfer protein TraQ | traQ |
| HPG17_RS00275 (BTU53_p074) | 48278..48991 | - | 714 | WP_000787003 | conjugal transfer protein TraP | traP |
| HPG17_RS00280 (BTU53_p075) | 48988..50325 | - | 1338 | WP_072105525 | conjugal transfer protein TraO | traO |
| HPG17_RS00285 (BTU53_p076) | 50329..51303 | - | 975 | WP_000547385 | DotH/IcmK family type IV secretion protein | traN |
| HPG17_RS00290 (BTU53_p077) | 51314..52009 | - | 696 | WP_001287545 | DotI/IcmL family type IV secretion protein | traM |
| HPG17_RS00295 (BTU53_p078) | 52021..52371 | - | 351 | WP_001056367 | hypothetical protein | traL |
| HPG17_RS00300 (BTU53_p079) | 52388..56449 | - | 4062 | WP_000842453 | LPD7 domain-containing protein | - |
| HPG17_RS00655 | 56513..56803 | - | 291 | WP_000817801 | IcmT/TraK family protein | traK |
| HPG17_RS00305 (BTU53_p081) | 56800..57948 | - | 1149 | WP_000775210 | plasmid transfer ATPase TraJ | virB11 |
| HPG17_RS00310 (BTU53_p082) | 57932..58768 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| HPG17_RS00315 (BTU53_p083) | 58765..59223 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| HPG17_RS00320 (BTU53_p085) | 59327..60529 | - | 1203 | WP_000979993 | conjugal transfer protein TraF | - |
| HPG17_RS00325 (BTU53_p086) | 60631..61452 | - | 822 | WP_000845373 | hypothetical protein | traE |
| HPG17_RS00670 | 61617..61811 | - | 195 | Protein_69 | integrase | - |
| HPG17_RS00330 (BTU53_p088) | 61925..63286 | - | 1362 | WP_001168171 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HPG17_RS00335 (BTU53_p089) | 63304..63840 | - | 537 | WP_023649457 | prepilin peptidase | - |
| HPG17_RS00340 (BTU53_p090) | 63946..64431 | - | 486 | WP_001080767 | lytic transglycosylase domain-containing protein | virB1 |
| HPG17_RS00345 (BTU53_p091) | 64476..65012 | - | 537 | WP_000111534 | type 4 pilus major pilin | - |
| HPG17_RS00350 (BTU53_p092) | 65074..65463 | - | 390 | Protein_74 | type II secretion system F family protein | - |
| HPG17_RS00355 | 65522..66216 | + | 695 | WP_087786203 | IS1-like element IS1N family transposase | - |
| HPG17_RS00360 (BTU53_p095) | 66232..66942 | - | 711 | Protein_76 | type II secretion system F family protein | - |
| HPG17_RS00365 (BTU53_p096) | 66944..68452 | - | 1509 | WP_032425678 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HPG17_RS00370 (BTU53_p097) | 68556..69014 | - | 459 | WP_000095885 | type IV pilus biogenesis protein PilP | - |
| HPG17_RS00375 (BTU53_p098) | 69004..70299 | - | 1296 | WP_000129883 | type 4b pilus protein PilO2 | - |
| HPG17_RS00380 (BTU53_p099) | 70320..71939 | - | 1620 | WP_001035565 | PilN family type IVB pilus formation outer membrane protein | - |
| HPG17_RS00385 (BTU53_p100) | 71971..72408 | - | 438 | WP_000539952 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 1073 | GenBank | NC_032099 |
| Plasmid name | p92-9000 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 121262 bp | Coordinate of oriT [Strand] | 24104..24189 [-] |
| Host baterium | Shigella dysenteriae |
Cargo genes
| Drug resistance gene | tet(B), catA1, ant(3'')-Ia, qacE, sul1 |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |