Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100612 |
| Name | oriT_pFI430 |
| Organism | Lactococcus cremoris subsp. cremoris MG1363 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_025249 (30906..30945 [+], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | 4..11, 16..23 (GTAAGATG..CATCTTAC) |
| Location of nic site | 31..32 |
| Conserved sequence flanking the nic site |
CTTG|CA |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 40 nt
>oriT_pFI430
TCCGTAAGATGCTATCATCTTACTATGCTTGCAAAAGGTC
TCCGTAAGATGCTATCATCTTACTATGCTTGCAAAAGGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file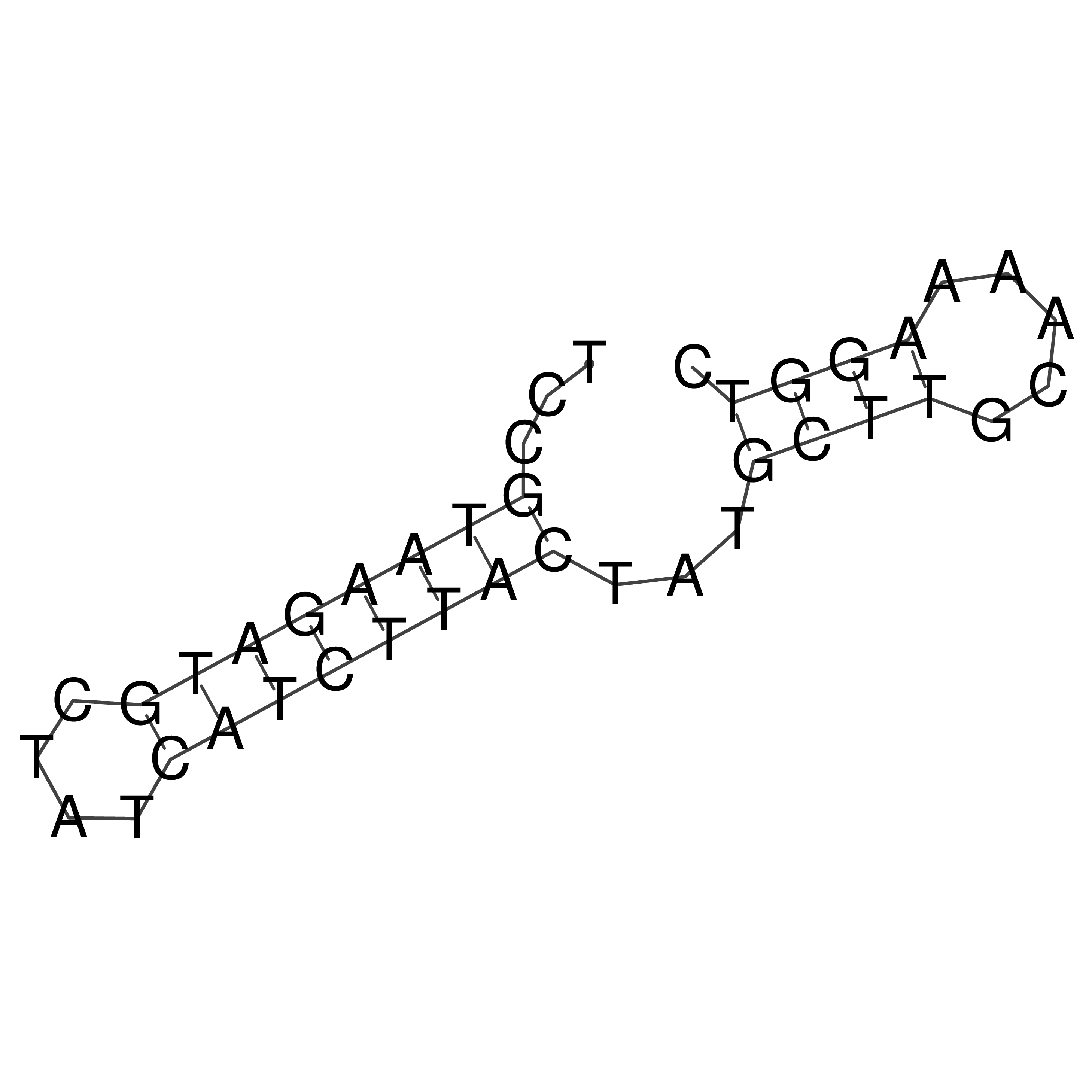
Relaxase
| ID | 684 | GenBank | YP_009091785 |
| Name | MobA_pFI430 |
UniProt ID | W5IDF4 |
| Length | 584 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 584 a.a. Molecular weight: 67146.02 Da Isoelectric Point: 6.5572
>YP_009091785.1 MobA (plasmid) [Lactococcus lactis subsp. cremoris MG1363]
MVYTKHIIVHKLKHLRQAKDYVENAEKTLVNESNEDHLTNLFPYISNPDKTMSKQLVSGHGITNVYDAAN
EFIATKKLKALSKGTDFNFDPQTGKVRFNVESLEKNNAVLGHHLIQSFSPDDNLTPEQIHEIGRQTILEF
TGGEYEFVIATHVDREHIHNVRPDRVLSQVVCEQGGTSLLHHIIFNSTNLYTGKQFDWKVIPKEKTKSGK
AYDVTKNNFEKVSDKIASRYGAKIIEKSPGNSHLKYTKWQTQSIYKSQIKQRLDYLLEMSSDIEDFKRKA
TALNLSFDFSGKWTTYRLLDEPQMKNTRGRNLDKNRPEKYNLESIIERLETNELSLTVDEVVERYEEKVD
VVKQDFDYQVTVEKGQIDHMTSKGFYLNVDFGIADRGQIFIGGYKVDQLENRDCVLYLKKNETFRLLSEK
EASFTKYLTGHDLAKQLGLYNGTVPLKKEPVISTINQLVDAINFLAEHGVTEGTQFNNMESQLMSALGEA
EEKLYVIDNKIMELTKIAKLLIEKESDHSQAVINELENLGVGPSIKYQDIHQELQSEKMSRKILKNKFEQ
TVDEINTFNEIRVTTLEENKGKIL
MVYTKHIIVHKLKHLRQAKDYVENAEKTLVNESNEDHLTNLFPYISNPDKTMSKQLVSGHGITNVYDAAN
EFIATKKLKALSKGTDFNFDPQTGKVRFNVESLEKNNAVLGHHLIQSFSPDDNLTPEQIHEIGRQTILEF
TGGEYEFVIATHVDREHIHNVRPDRVLSQVVCEQGGTSLLHHIIFNSTNLYTGKQFDWKVIPKEKTKSGK
AYDVTKNNFEKVSDKIASRYGAKIIEKSPGNSHLKYTKWQTQSIYKSQIKQRLDYLLEMSSDIEDFKRKA
TALNLSFDFSGKWTTYRLLDEPQMKNTRGRNLDKNRPEKYNLESIIERLETNELSLTVDEVVERYEEKVD
VVKQDFDYQVTVEKGQIDHMTSKGFYLNVDFGIADRGQIFIGGYKVDQLENRDCVLYLKKNETFRLLSEK
EASFTKYLTGHDLAKQLGLYNGTVPLKKEPVISTINQLVDAINFLAEHGVTEGTQFNNMESQLMSALGEA
EEKLYVIDNKIMELTKIAKLLIEKESDHSQAVINELENLGVGPSIKYQDIHQELQSEKMSRKILKNKFEQ
TVDEINTFNEIRVTTLEENKGKIL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W5IDF4 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 7600..23649
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HS325_RS00020 (D688_p3004) | 2999..3196 | + | 198 | WP_011835268 | hypothetical protein | - |
| HS325_RS00025 (D688_p3005) | 3210..3407 | + | 198 | WP_011835267 | hypothetical protein | - |
| HS325_RS00030 (D688_p3006) | 3412..3675 | + | 264 | WP_011835266 | hypothetical protein | - |
| HS325_RS00035 | 3677..3820 | + | 144 | WP_014735049 | hypothetical protein | - |
| HS325_RS00040 | 3836..4081 | + | 246 | WP_014735048 | hypothetical protein | - |
| HS325_RS00045 (D688_p3007) | 4086..4625 | + | 540 | WP_011835265 | hypothetical protein | - |
| HS325_RS00050 (D688_p3008) | 5055..5417 | + | 363 | WP_011835264 | hypothetical protein | - |
| HS325_RS00055 | 5572..5805 | + | 234 | WP_014735047 | hypothetical protein | - |
| HS325_RS00060 (D688_p3009) | 6051..6350 | + | 300 | WP_011835263 | DnaJ domain-containing protein | - |
| HS325_RS00065 (D688_p3010) | 6368..7390 | + | 1023 | WP_011835262 | hypothetical protein | - |
| HS325_RS00070 (D688_p3011) | 7600..11331 | + | 3732 | WP_011835261 | SspB-related isopeptide-forming adhesin | prgB |
| HS325_RS00075 (D688_p3014) | 11702..12100 | + | 399 | WP_011835258 | hypothetical protein | - |
| HS325_RS00080 | 12152..12379 | + | 228 | WP_014735045 | hypothetical protein | - |
| HS325_RS00085 (D688_p3015) | 12391..13179 | + | 789 | WP_011835257 | hypothetical protein | prgHb |
| HS325_RS00090 (D688_p3016) | 13181..13534 | + | 354 | WP_011835256 | hypothetical protein | prgIc |
| HS325_RS00095 (D688_p3017) | 13488..15878 | + | 2391 | WP_041931300 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| HS325_RS00100 (D688_p3018) | 15882..18494 | + | 2613 | WP_011835254 | phage tail tip lysozyme | prgK |
| HS325_RS00105 (D688_p3019) | 18504..18821 | + | 318 | WP_011835253 | hypothetical protein | - |
| HS325_RS00110 (D688_p3020) | 18847..19539 | + | 693 | WP_011835252 | hypothetical protein | prgL |
| HS325_RS00115 (D688_p3021) | 19540..19785 | + | 246 | WP_011835251 | hypothetical protein | - |
| HS325_RS00120 (D688_p3022) | 19785..21098 | + | 1314 | WP_011835250 | hypothetical protein | - |
| HS325_RS00125 (D688_p3023) | 21114..21332 | + | 219 | WP_011835249 | hypothetical protein | - |
| HS325_RS00130 (D688_p3024) | 21311..21814 | + | 504 | WP_011835248 | hypothetical protein | - |
| HS325_RS00135 (D688_p3025) | 21811..23649 | + | 1839 | WP_011835247 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| HS325_RS00140 (D688_p3026) | 23675..24739 | + | 1065 | WP_011835246 | hypothetical protein | - |
| HS325_RS00145 (D688_p3027) | 24848..25042 | + | 195 | WP_011835245 | hypothetical protein | - |
| HS325_RS00150 (D688_p3028) | 25104..26288 | + | 1185 | WP_011835244 | toprim domain-containing protein | - |
| HS325_RS00155 (D688_p3029) | 26302..26817 | + | 516 | WP_011835243 | hypothetical protein | - |
| HS325_RS00160 (D688_p3030) | 26798..28564 | + | 1767 | WP_011835242 | hypothetical protein | - |
Host bacterium
| ID | 1072 | GenBank | NC_025249 |
| Plasmid name | pFI430 | Incompatibility group | _ |
| Plasmid size | 59474 bp | Coordinate of oriT [Strand] | 30906..30945 [+] |
| Host baterium | Lactococcus cremoris subsp. cremoris MG1363 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |