Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100601 |
| Name | oriT_ColicinE1 |
| Organism | Escherichia coli |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_025157 (93..175 [+], 83 nt) |
| oriT length | 83 nt |
| IRs (inverted repeats) | 3..12, 16..25 (GTGTCGGGGC..GCCCTGACCC) |
| Location of nic site | 56..57 |
| Conserved sequence flanking the nic site |
CTGG|CTTA |
| Note | predicted by the oriTfinder |
oriT sequence
Download Length: 83 nt
>oriT_ColicinE1
GGGTGTCGGGGCGCAGCCCTGACCCAGTCACGTAGCGATAGCGGAGTGTATACTGGCTTAACCATGCGGCATCAGTGCGGATT
GGGTGTCGGGGCGCAGCCCTGACCCAGTCACGTAGCGATAGCGGAGTGTATACTGGCTTAACCATGCGGCATCAGTGCGGATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file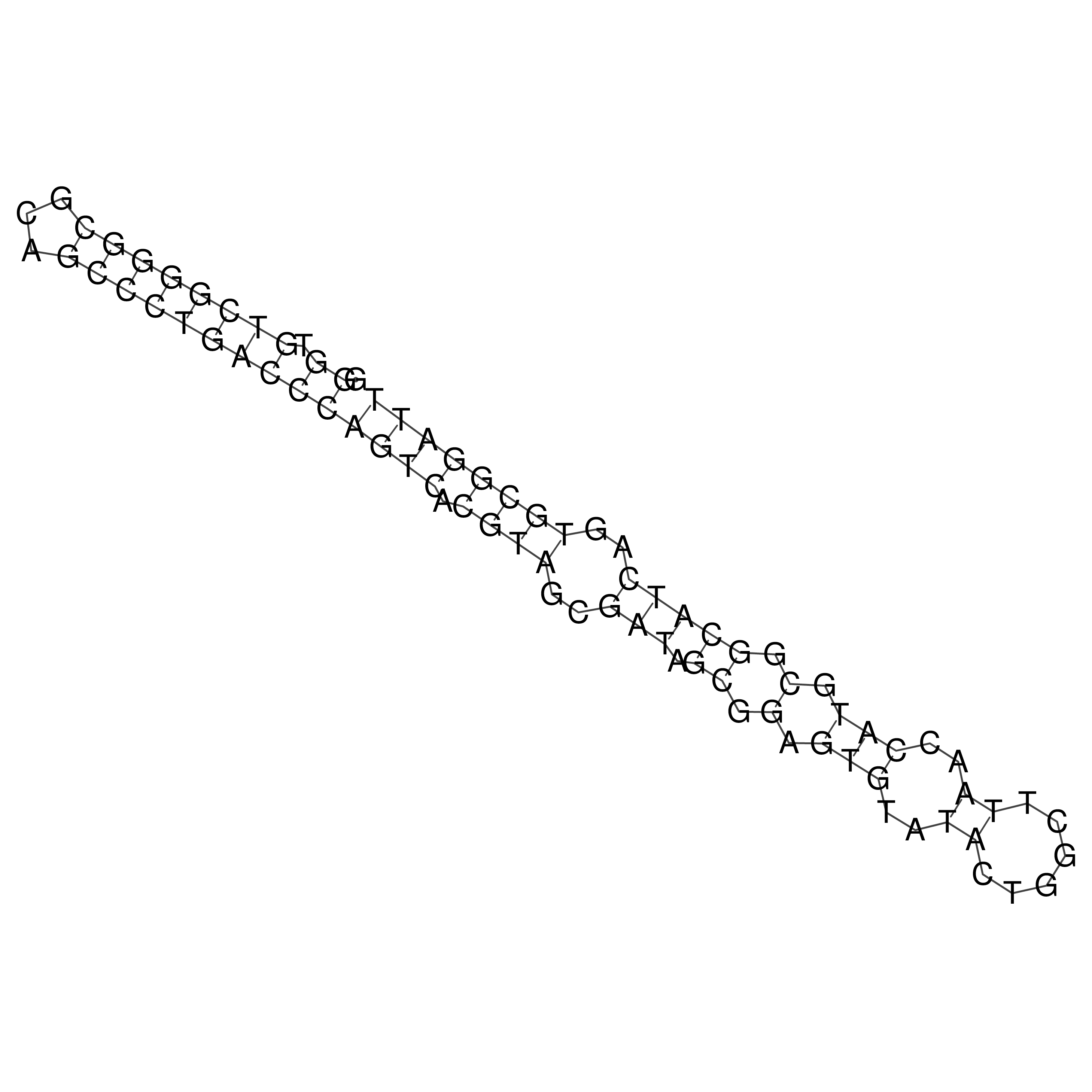
Relaxase
| ID | 673 | GenBank | YP_009070583 |
| Name | TraI_ColicinE1 |
UniProt ID | A0A075M947 |
| Length | 1763 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 1763 a.a. Molecular weight: 193017.09 Da Isoelectric Point: 5.9894
>YP_009070583.1 IncF plasmid conjugative transfer DNA-nicking and unwinding protein TraI (plasmid) [Escherichia coli]
MSKGYTFMMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDG
ADLSRMQDGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSET
VLTGNLVMALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREK
LKEQVEALGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEIK
MVEWMQTLKETGFDIRAYRDAADQRADLRTLTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLART
VGILPPENGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSV
PRTAGYSDAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNMKQDERLS
GELITGRRQLLEGMTFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLIIDSGQRTGTGSALMAMKDA
GVNTYRWQGGEQRPATIISEPDRNVRYARLAGDFAVSVKAGEESVAQVSGVREQAMLTQTIRSELKTQGV
LGHPEVTMTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVV
RISSLDSSWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVSSVSEDAMTVVVPGRAEPATLPVSDS
PFTALKLENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSF
TVVSEQIKTRAGETSLETAISHQKSALHTPAQQAIHLALPVVESKKLAFSMVDLLTEAKSFAAEGTGFTE
LGGEINAQIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVMPLMERVPGELMEKLTSGQ
RAATRMILETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTL
ASFLHDTQLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFR
LQQTRSAADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQ
EAKLAEAQQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGE
LGKEQVMVPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLQDAEGNTRLISPRE
AVAEGVTLYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTREIRPGQEQAEQH
IDLAYAITAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQK
GTAHDVFEPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFD
RNGKSAGIWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGV
VVRIAGEGRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMA
ENKPDLPDGKTEQAVREIAGQERDRAAITEREAALPEGVLREPQRVREAVREIARENLLQERLQQMERDM
VRDLQKEKTLGGD
MSKGYTFMMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDG
ADLSRMQDGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSET
VLTGNLVMALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREK
LKEQVEALGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEIK
MVEWMQTLKETGFDIRAYRDAADQRADLRTLTPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLART
VGILPPENGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSV
PRTAGYSDAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNMKQDERLS
GELITGRRQLLEGMTFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLIIDSGQRTGTGSALMAMKDA
GVNTYRWQGGEQRPATIISEPDRNVRYARLAGDFAVSVKAGEESVAQVSGVREQAMLTQTIRSELKTQGV
LGHPEVTMTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVV
RISSLDSSWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVSSVSEDAMTVVVPGRAEPATLPVSDS
PFTALKLENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSF
TVVSEQIKTRAGETSLETAISHQKSALHTPAQQAIHLALPVVESKKLAFSMVDLLTEAKSFAAEGTGFTE
LGGEINAQIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVMPLMERVPGELMEKLTSGQ
RAATRMILETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTL
ASFLHDTQLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFR
LQQTRSAADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQ
EAKLAEAQQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGE
LGKEQVMVPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLQDAEGNTRLISPRE
AVAEGVTLYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTREIRPGQEQAEQH
IDLAYAITAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQK
GTAHDVFEPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFD
RNGKSAGIWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGV
VVRIAGEGRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMA
ENKPDLPDGKTEQAVREIAGQERDRAAITEREAALPEGVLREPQRVREAVREIARENLLQERLQQMERDM
VRDLQKEKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 1061 | GenBank | NC_025157 |
| Plasmid name | Colicin E1 | Incompatibility group | ColRNAI |
| Plasmid size | 1683 bp | Coordinate of oriT [Strand] | 93..175 [+] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |